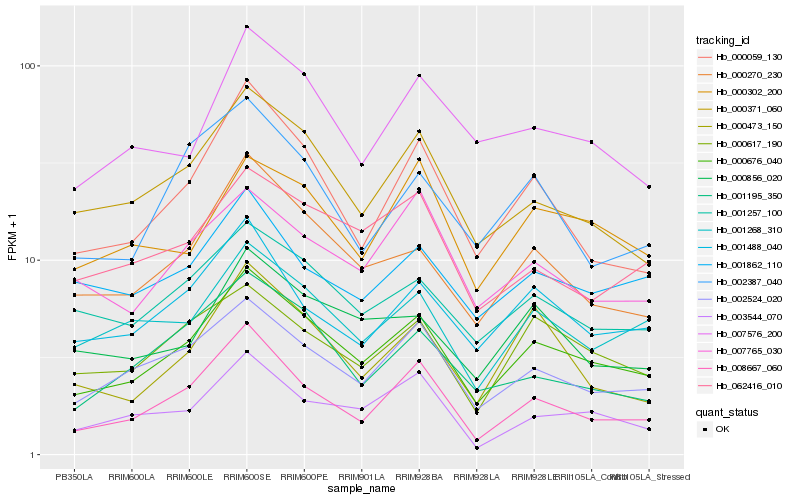
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 2 |
Hb_002524_020 |
0.0879015597 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 3 |
Hb_000270_230 |
0.0909506728 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 4 |
Hb_000371_060 |
0.0966046629 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 5 |
Hb_001257_100 |
0.0989748382 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_008667_060 |
0.0994074505 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 7 |
Hb_000059_130 |
0.1002744767 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 8 |
Hb_001268_310 |
0.1019577143 |
- |
- |
- |
| 9 |
Hb_000617_190 |
0.1029238538 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 10 |
Hb_001488_040 |
0.1039802719 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 11 |
Hb_000856_020 |
0.1111251143 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 12 |
Hb_007576_200 |
0.1114980327 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_003544_070 |
0.1150618618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_062416_010 |
0.1160610746 |
- |
- |
PREDICTED: UPF0202 protein At1g10490 [Jatropha curcas] |
| 15 |
Hb_000302_200 |
0.118016012 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 16 |
Hb_000473_150 |
0.1183429256 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007765_030 |
0.1202197595 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 18 |
Hb_001195_350 |
0.1207949611 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 19 |
Hb_001862_110 |
0.1213819261 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_002387_040 |
0.1214692568 |
- |
- |
protein transporter, putative [Ricinus communis] |