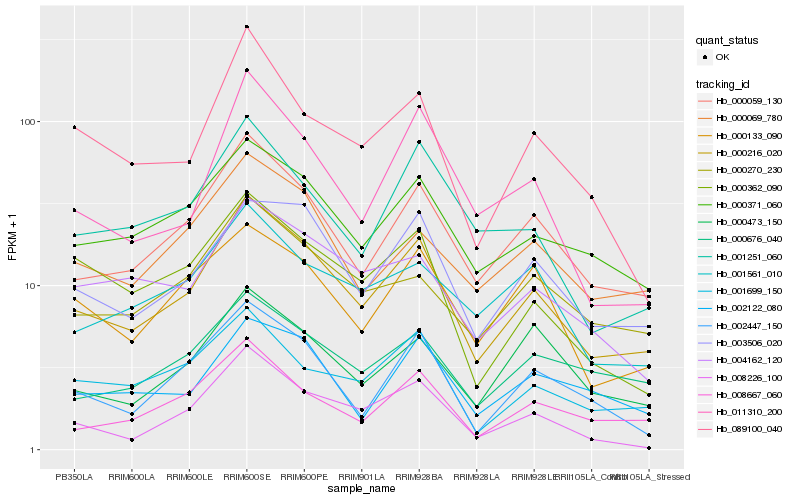
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000216_020 |
0.0 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 2 |
Hb_000059_130 |
0.0799712977 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 3 |
Hb_003506_020 |
0.1018418165 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 4 |
Hb_000371_060 |
0.1031332829 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000270_230 |
0.1168839971 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 6 |
Hb_008667_060 |
0.1177389057 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 7 |
Hb_000069_780 |
0.1182244628 |
- |
- |
atpob1, putative [Ricinus communis] |
| 8 |
Hb_002447_150 |
0.1200612763 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 9 |
Hb_001699_150 |
0.1218768091 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 10 |
Hb_000133_090 |
0.1237430059 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 11 |
Hb_011310_200 |
0.1237658225 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 12 |
Hb_008226_100 |
0.1239532792 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_089100_040 |
0.1254398237 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 14 |
Hb_000676_040 |
0.1265164429 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 15 |
Hb_000473_150 |
0.1272481083 |
- |
- |
PREDICTED: pullulanase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004162_120 |
0.127387957 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 17 |
Hb_001251_060 |
0.1280843479 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 18 |
Hb_000362_090 |
0.1297185268 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001561_010 |
0.1304936411 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002122_080 |
0.1305119644 |
- |
- |
- |