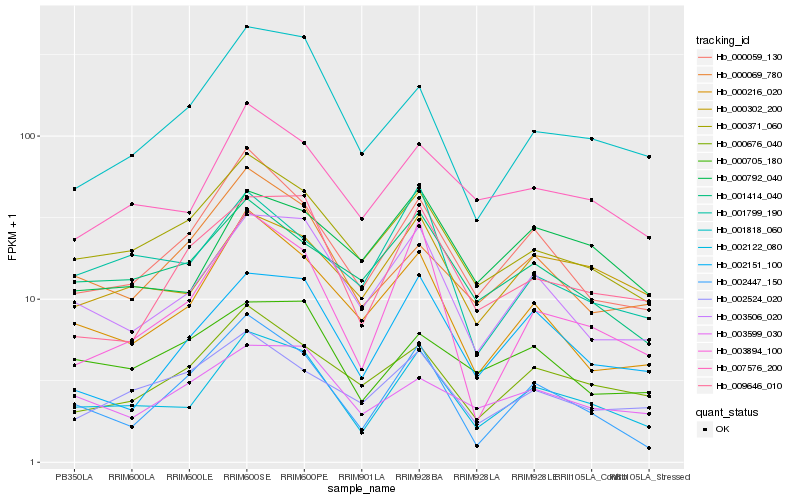
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002122_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_003506_020 |
0.1047032984 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 3 |
Hb_007576_200 |
0.113575959 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_000371_060 |
0.1148470964 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000302_200 |
0.1178798981 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 6 |
Hb_000059_130 |
0.1213554884 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000216_020 |
0.1305119644 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 8 |
Hb_001414_040 |
0.1345308832 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002151_100 |
0.135492798 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 10 |
Hb_000792_040 |
0.1365608895 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 11 |
Hb_000676_040 |
0.137157609 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 12 |
Hb_003894_100 |
0.1380545881 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 13 |
Hb_002447_150 |
0.1404588984 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_001799_190 |
0.1438402232 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 15 |
Hb_009646_010 |
0.1445730915 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 16 |
Hb_000705_180 |
0.1459639233 |
- |
- |
hypothetical protein JCGZ_02923 [Jatropha curcas] |
| 17 |
Hb_001818_060 |
0.1466285154 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 18 |
Hb_000069_780 |
0.148191061 |
- |
- |
atpob1, putative [Ricinus communis] |
| 19 |
Hb_003599_030 |
0.149399255 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_002524_020 |
0.1496372687 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |