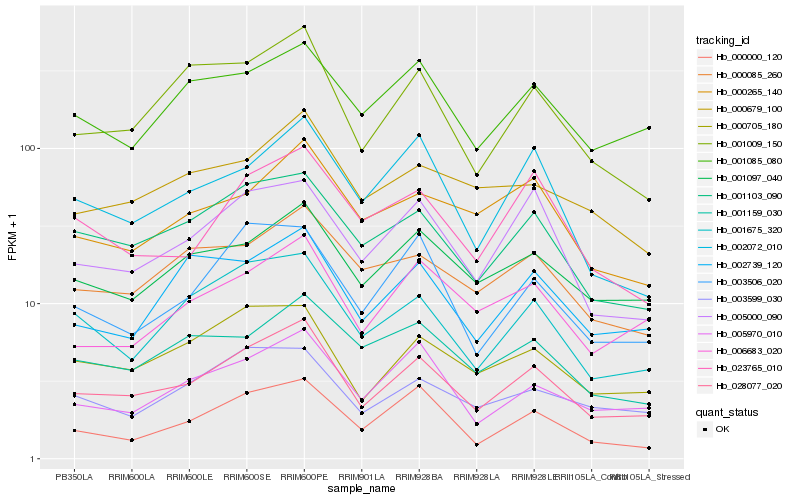
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028077_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001103_090 |
0.0907706412 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 3 |
Hb_001675_320 |
0.0991745906 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 4 |
Hb_000705_180 |
0.0993061386 |
- |
- |
hypothetical protein JCGZ_02923 [Jatropha curcas] |
| 5 |
Hb_001009_150 |
0.1010162531 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 6 |
Hb_001097_040 |
0.1011127494 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 7 |
Hb_005970_010 |
0.1039453793 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005000_090 |
0.1054375074 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 9 |
Hb_006683_020 |
0.1100298628 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 10 |
Hb_000000_120 |
0.1112735351 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 11 |
Hb_001085_080 |
0.1132236536 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 12 |
Hb_002739_120 |
0.1144089642 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 13 |
Hb_003506_020 |
0.1144637426 |
- |
- |
PREDICTED: uncharacterized protein LOC105125881 isoform X1 [Populus euphratica] |
| 14 |
Hb_000085_260 |
0.1147213092 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 15 |
Hb_003599_030 |
0.1155472484 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_023765_010 |
0.1160793125 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 17 |
Hb_002072_010 |
0.1176372088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000265_140 |
0.1192841499 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 19 |
Hb_001159_030 |
0.1196563467 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 20 |
Hb_000679_100 |
0.119898924 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |