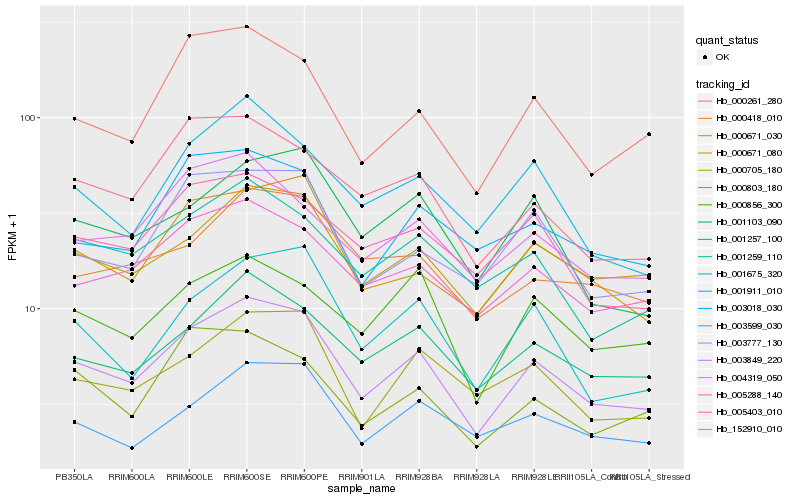
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003849_220 |
0.0 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 2 |
Hb_000671_080 |
0.0827828209 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 3 |
Hb_001257_100 |
0.0866144181 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 4 |
Hb_000261_280 |
0.0892943631 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 5 |
Hb_000671_030 |
0.0898110242 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 6 |
Hb_005288_140 |
0.0909822893 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 7 |
Hb_003777_130 |
0.0962375452 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 8 |
Hb_001259_110 |
0.0976684101 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 9 |
Hb_001675_320 |
0.0979617767 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 10 |
Hb_001911_010 |
0.0981949451 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 11 |
Hb_152910_010 |
0.0983439393 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 12 |
Hb_000705_180 |
0.099644525 |
- |
- |
hypothetical protein JCGZ_02923 [Jatropha curcas] |
| 13 |
Hb_003599_030 |
0.0998129707 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 14 |
Hb_004319_050 |
0.1006845207 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 15 |
Hb_005403_010 |
0.1012357164 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 16 |
Hb_003018_030 |
0.102816399 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 17 |
Hb_000856_300 |
0.1030951599 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |
| 18 |
Hb_001103_090 |
0.1051763666 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 19 |
Hb_000803_180 |
0.1063589008 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_000418_010 |
0.1072303418 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |