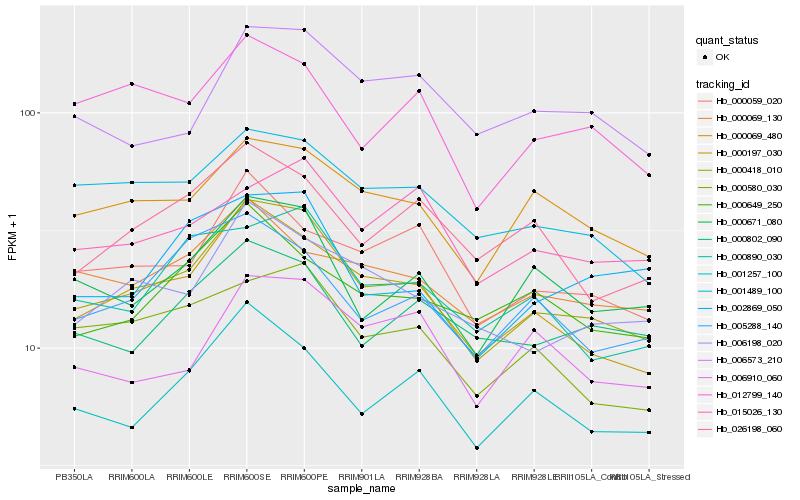
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000418_010 |
0.0 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 2 |
Hb_000197_030 |
0.063386548 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 3 |
Hb_000069_480 |
0.0800401035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000671_080 |
0.080540247 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 5 |
Hb_001257_100 |
0.0851633357 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_001489_100 |
0.0857868475 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 7 |
Hb_006198_020 |
0.0888590112 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 8 |
Hb_000580_030 |
0.090754178 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 9 |
Hb_000059_020 |
0.0928612326 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 10 |
Hb_000890_030 |
0.0930242532 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 11 |
Hb_005288_140 |
0.0964033428 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 12 |
Hb_012799_140 |
0.0966694812 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 13 |
Hb_000802_090 |
0.0977148884 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 14 |
Hb_026198_060 |
0.0985466425 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 15 |
Hb_000649_250 |
0.0993321179 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_002869_050 |
0.0997681442 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_000069_130 |
0.0997886125 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006573_210 |
0.1008531173 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 19 |
Hb_006910_060 |
0.1029333879 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780-like [Jatropha curcas] |
| 20 |
Hb_015026_130 |
0.1037999619 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |