| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
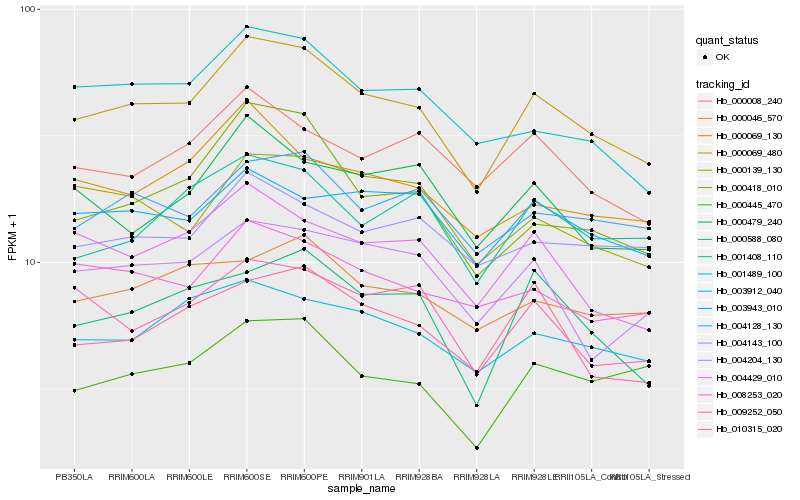
Hb_000069_480 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003912_040 |
0.071385694 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 3 |
Hb_004128_130 |
0.075372209 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004429_010 |
0.0784472512 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_001489_100 |
0.0791904768 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 6 |
Hb_008253_020 |
0.0794495706 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_000418_010 |
0.0800401035 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 8 |
Hb_000139_130 |
0.0804789796 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 9 |
Hb_010315_020 |
0.0809608681 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 10 |
Hb_000588_080 |
0.0811167062 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 11 |
Hb_001408_110 |
0.081157696 |
- |
- |
hypothetical protein EUGRSUZ_I02762 [Eucalyptus grandis] |
| 12 |
Hb_009252_050 |
0.0811729032 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000069_130 |
0.0811810358 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003943_010 |
0.0815629832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000008_240 |
0.0819384009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000445_470 |
0.0829002791 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_000046_570 |
0.0838724221 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 18 |
Hb_004204_130 |
0.0843139798 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 19 |
Hb_004143_100 |
0.0845911379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000479_240 |
0.0851172265 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |