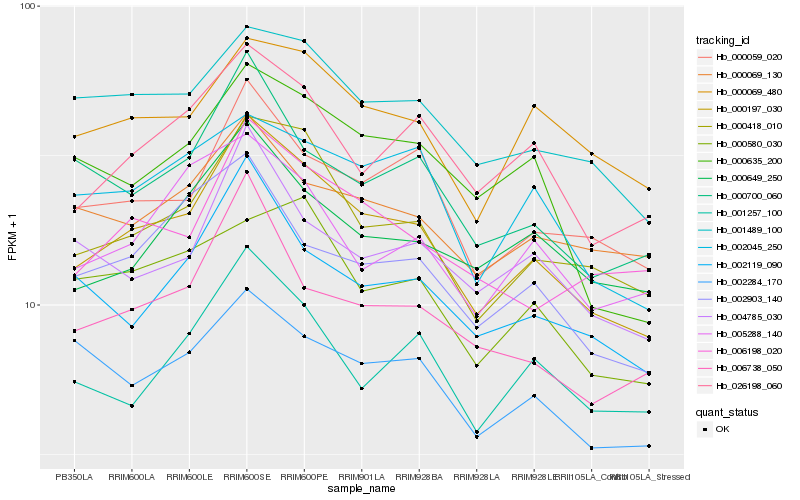
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_030 |
0.0 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 2 |
Hb_000418_010 |
0.063386548 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 3 |
Hb_000059_020 |
0.0770321923 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 4 |
Hb_001489_100 |
0.0813117514 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 5 |
Hb_026198_060 |
0.0850533692 |
- |
- |
PREDICTED: calcium-dependent protein kinase 11 [Jatropha curcas] |
| 6 |
Hb_002119_090 |
0.0866955339 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 7 |
Hb_006198_020 |
0.0877479758 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 8 |
Hb_000700_060 |
0.0880189821 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 9 |
Hb_006738_050 |
0.0891626261 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 10 |
Hb_000069_480 |
0.0897083494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000649_250 |
0.0901059914 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 12 |
Hb_002045_250 |
0.0908184248 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 13 |
Hb_001257_100 |
0.0911080851 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 14 |
Hb_002903_140 |
0.0912090968 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 15 |
Hb_000635_200 |
0.0920441183 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 16 |
Hb_004785_030 |
0.0924691038 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 17 |
Hb_002284_170 |
0.0934639315 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 18 |
Hb_000069_130 |
0.0945194485 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000580_030 |
0.0959168233 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 20 |
Hb_005288_140 |
0.0970331032 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |