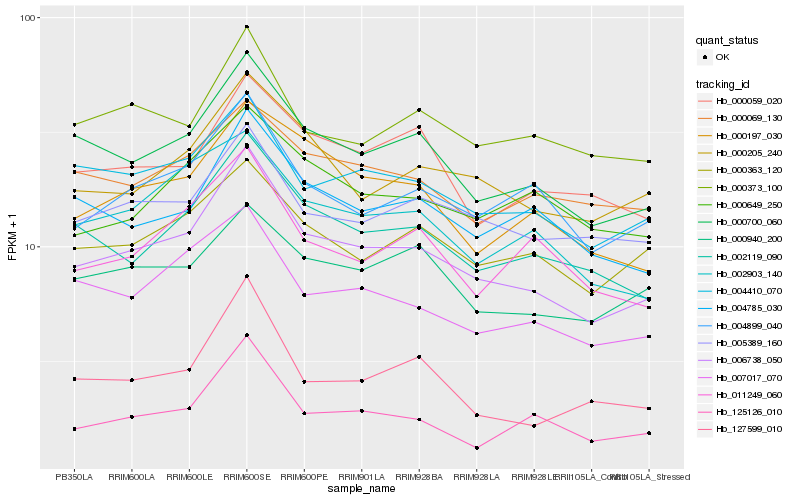
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006738_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 2 |
Hb_000700_060 |
0.0622698027 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 3 |
Hb_002119_090 |
0.0752897336 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 4 |
Hb_004410_070 |
0.0768865545 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 5 |
Hb_000205_240 |
0.0827552419 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_007017_070 |
0.0829240247 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004899_040 |
0.0849459361 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 8 |
Hb_004785_030 |
0.0856222628 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 9 |
Hb_000197_030 |
0.0891626261 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 10 |
Hb_005389_160 |
0.0897408739 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 11 |
Hb_002903_140 |
0.0905827975 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 12 |
Hb_125126_010 |
0.0929057147 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 13 |
Hb_000363_120 |
0.0934677492 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 14 |
Hb_000373_100 |
0.0957560014 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 15 |
Hb_011249_060 |
0.0958409587 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 16 |
Hb_127599_010 |
0.0969110683 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 17 |
Hb_000059_020 |
0.0971352427 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 18 |
Hb_000940_200 |
0.098800246 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 19 |
Hb_000649_250 |
0.0998961576 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_000069_130 |
0.1003195955 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |