| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
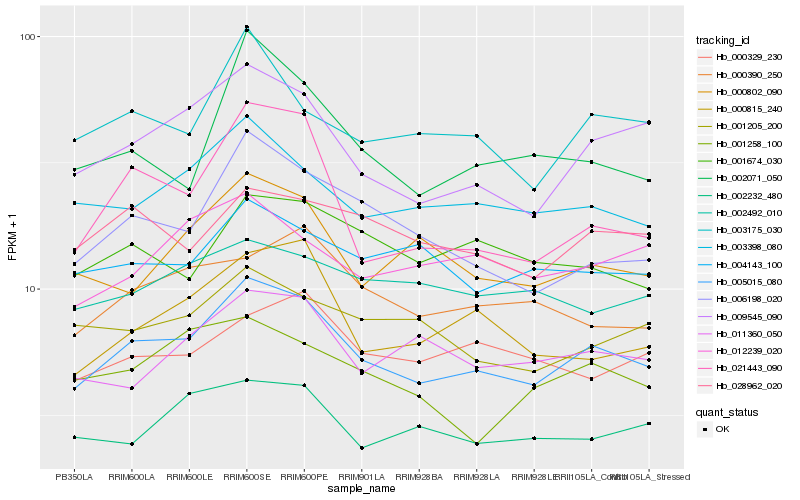
Hb_005015_080 |
0.0 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_009545_090 |
0.0750341717 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 3 |
Hb_003398_080 |
0.086107536 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_006198_020 |
0.087965013 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 5 |
Hb_003175_030 |
0.093028429 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 6 |
Hb_000815_240 |
0.0955420811 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_021443_090 |
0.0963405044 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 8 |
Hb_000802_090 |
0.0972039358 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_000390_250 |
0.0988040798 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 10 |
Hb_011360_050 |
0.0988897197 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 11 |
Hb_028962_020 |
0.0991001933 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 12 |
Hb_002232_480 |
0.1001541189 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 13 |
Hb_002071_050 |
0.1013792559 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 14 |
Hb_004143_100 |
0.1018147565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001674_030 |
0.1021526048 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 16 |
Hb_001205_200 |
0.1023629836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_012239_020 |
0.1024673841 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 18 |
Hb_000329_230 |
0.1053389528 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 19 |
Hb_001258_100 |
0.1063709227 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_002492_010 |
0.1069741345 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |