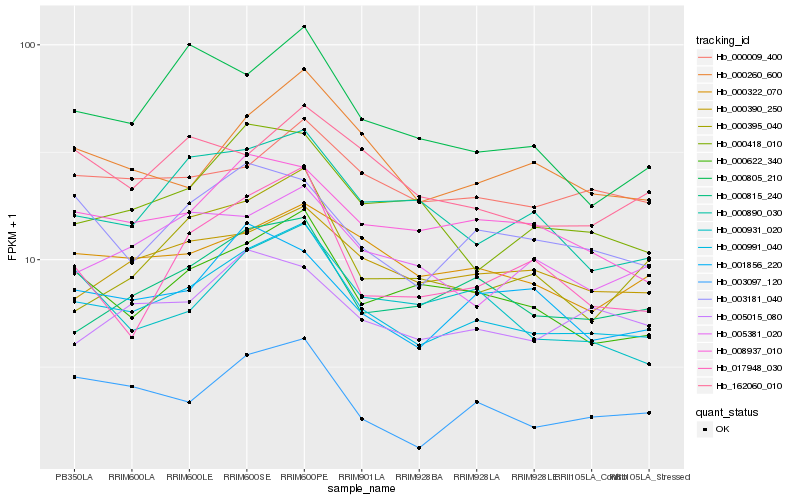
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000991_040 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 2 |
Hb_000622_340 |
0.0913481224 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 3 |
Hb_000260_600 |
0.0937730999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000815_240 |
0.0951433019 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_008937_010 |
0.1015532399 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000390_250 |
0.1053160171 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 7 |
Hb_000322_070 |
0.1063955988 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 8 |
Hb_000890_030 |
0.1064466382 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 9 |
Hb_003181_040 |
0.1073993215 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003097_120 |
0.1075570077 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 11 |
Hb_000418_010 |
0.1076508465 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 12 |
Hb_000931_020 |
0.1097423503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005381_020 |
0.109855956 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 14 |
Hb_017948_030 |
0.1100013288 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 15 |
Hb_000009_400 |
0.110436473 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 16 |
Hb_001856_220 |
0.1119087012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000395_040 |
0.1129536257 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 18 |
Hb_162060_010 |
0.114332402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005015_080 |
0.1143934953 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000805_210 |
0.1145888164 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |