| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
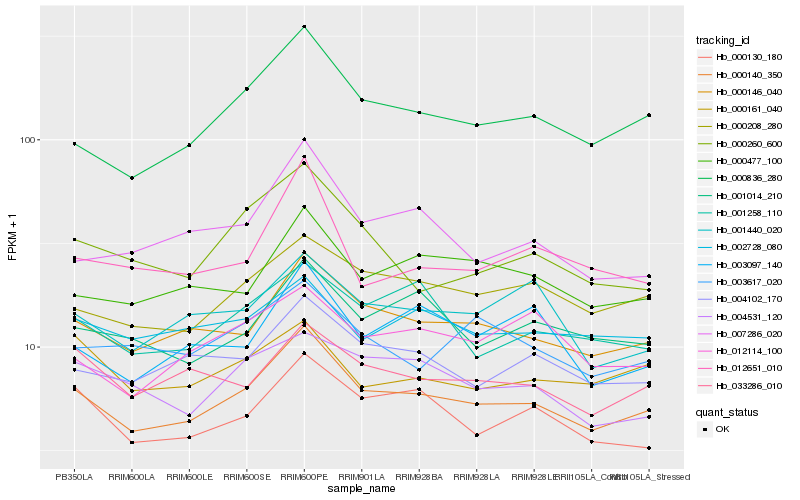
Hb_000140_350 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000146_040 |
0.056866387 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 3 |
Hb_000836_280 |
0.0671396542 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000208_280 |
0.0757223881 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 5 |
Hb_004102_170 |
0.0782046834 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 6 |
Hb_000130_180 |
0.0797543766 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000477_100 |
0.0837064595 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 8 |
Hb_001014_210 |
0.0854774743 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 9 |
Hb_033286_010 |
0.087562559 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 10 |
Hb_001258_110 |
0.090995522 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_001440_020 |
0.092715586 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 12 |
Hb_002728_080 |
0.0928952651 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_007286_020 |
0.093796659 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 14 |
Hb_012114_100 |
0.095799738 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_003097_140 |
0.0958660278 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 16 |
Hb_004531_120 |
0.0959659023 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 17 |
Hb_003617_020 |
0.0966163258 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 18 |
Hb_000260_600 |
0.0972292744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012651_010 |
0.0982218336 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 20 |
Hb_000161_040 |
0.0987983993 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |