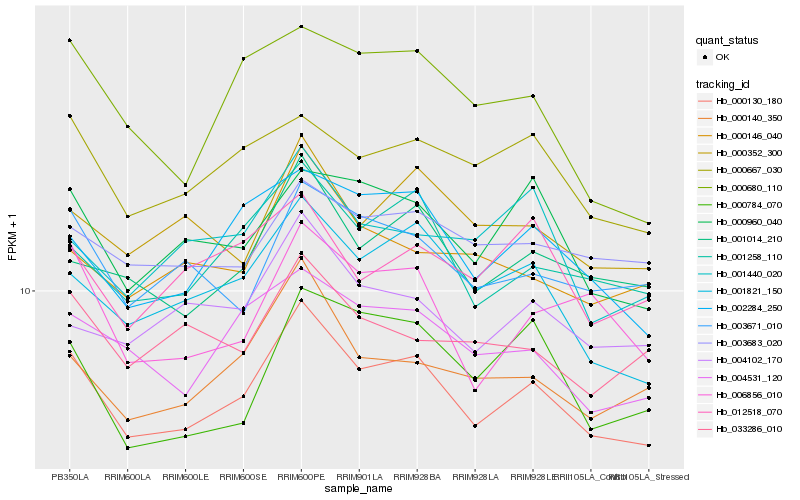
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_180 |
0.0 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001258_110 |
0.0727660518 |
- |
- |
transporter, putative [Ricinus communis] |
| 3 |
Hb_001821_150 |
0.0744616036 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 4 |
Hb_001014_210 |
0.0774623006 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 5 |
Hb_000140_350 |
0.0797543766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002284_250 |
0.0810978053 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 7 |
Hb_004531_120 |
0.0812666315 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 8 |
Hb_000960_040 |
0.0851640969 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 9 |
Hb_003671_010 |
0.0865445305 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006856_010 |
0.0868942748 |
- |
- |
hypothetical protein CISIN_1g019592mg [Citrus sinensis] |
| 11 |
Hb_000667_030 |
0.0883618294 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 12 |
Hb_000352_300 |
0.090961869 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 13 |
Hb_004102_170 |
0.0914219998 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 14 |
Hb_000146_040 |
0.0925447063 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 15 |
Hb_003683_020 |
0.0956016785 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000680_110 |
0.0957423706 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 17 |
Hb_033286_010 |
0.0978567474 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 18 |
Hb_012518_070 |
0.0978885434 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001440_020 |
0.098305293 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 20 |
Hb_000784_070 |
0.0986185022 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |