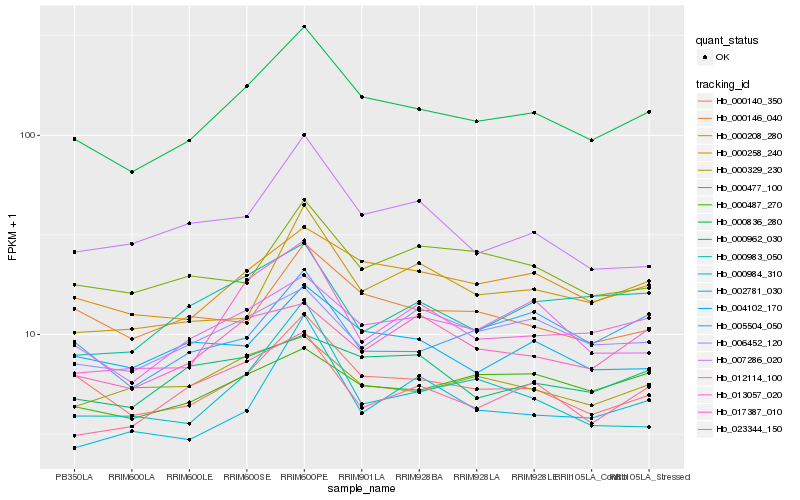
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_280 |
0.0 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000140_350 |
0.0671396542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000208_280 |
0.077741211 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 4 |
Hb_013057_020 |
0.0869248545 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 5 |
Hb_023344_150 |
0.0933469499 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 6 |
Hb_012114_100 |
0.0943085223 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 7 |
Hb_000146_040 |
0.0962883656 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 8 |
Hb_004102_170 |
0.0965320221 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 9 |
Hb_000258_240 |
0.1000328376 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 10 |
Hb_007286_020 |
0.1004615822 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 11 |
Hb_000984_310 |
0.1027811022 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 12 |
Hb_005504_050 |
0.1041215723 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 13 |
Hb_000477_100 |
0.1052878463 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 14 |
Hb_000329_230 |
0.1082022868 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 15 |
Hb_006452_120 |
0.1082086068 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 16 |
Hb_017387_010 |
0.1085782582 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 17 |
Hb_000487_270 |
0.1087609085 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 18 |
Hb_000983_050 |
0.1094283866 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_002781_030 |
0.1096758364 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000962_030 |
0.1106021285 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |