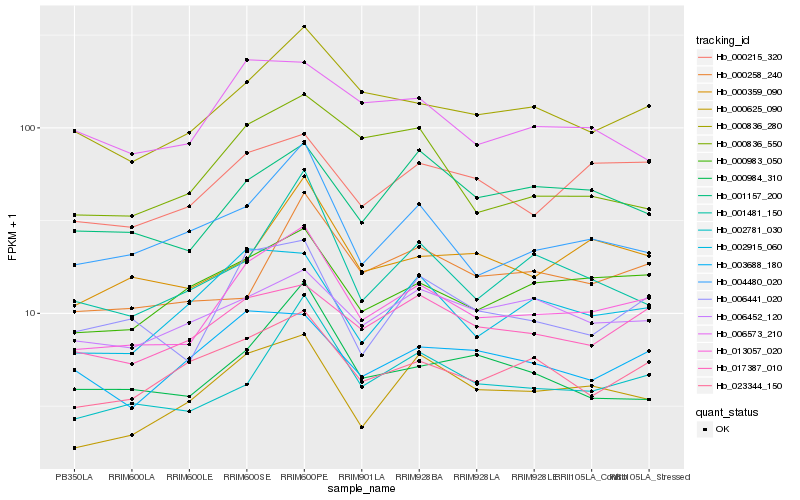
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013057_020 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 2 |
Hb_000836_280 |
0.0869248545 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_006441_020 |
0.0922682542 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 4 |
Hb_000983_050 |
0.097127653 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_023344_150 |
0.1021020057 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 6 |
Hb_002781_030 |
0.1028039554 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_004480_020 |
0.1032322852 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 8 |
Hb_000625_090 |
0.1059913858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000215_320 |
0.108871229 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 10 |
Hb_000359_090 |
0.1096124055 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 11 |
Hb_017387_010 |
0.110725936 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 12 |
Hb_002915_060 |
0.1120197876 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 13 |
Hb_001157_200 |
0.1170705075 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 14 |
Hb_000984_310 |
0.1197136877 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 15 |
Hb_003688_180 |
0.120715711 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 16 |
Hb_001481_150 |
0.1224217906 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 17 |
Hb_006452_120 |
0.1224958246 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 18 |
Hb_006573_210 |
0.1241629896 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 19 |
Hb_000836_550 |
0.124298503 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 20 |
Hb_000258_240 |
0.1244209884 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |