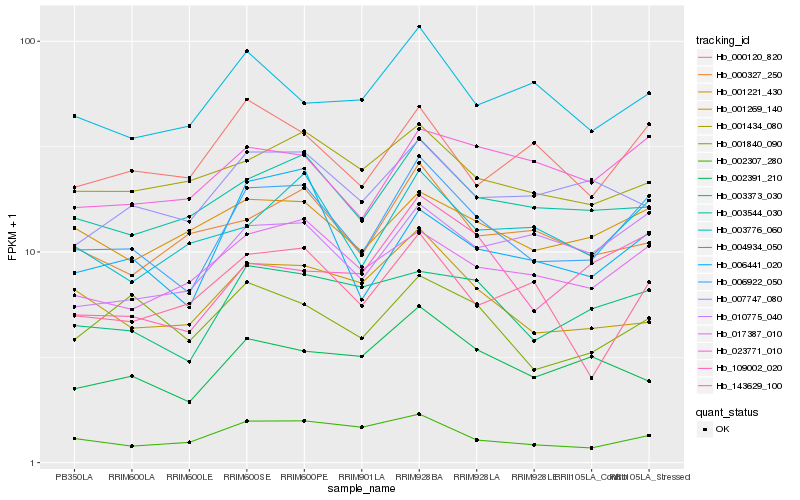
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006922_050 |
0.0 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 2 |
Hb_003544_030 |
0.1049968199 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 3 |
Hb_017387_010 |
0.1073428144 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 4 |
Hb_003373_030 |
0.1084652049 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATG1 [Jatropha curcas] |
| 5 |
Hb_003776_060 |
0.1134848481 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 6 |
Hb_010775_040 |
0.1175437476 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 7 |
Hb_023771_010 |
0.117749915 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001221_430 |
0.1190122303 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At5g63520 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000120_820 |
0.1190950886 |
- |
- |
dihydrodipicolinate synthase, putative [Ricinus communis] |
| 10 |
Hb_002307_280 |
0.1194736569 |
- |
- |
Endonuclease/exonuclease/phosphatase family protein isoform 3 [Theobroma cacao] |
| 11 |
Hb_001840_090 |
0.1206323925 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_006441_020 |
0.1206570778 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 13 |
Hb_001269_140 |
0.1227837853 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 14 |
Hb_001434_080 |
0.1236650082 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 15 |
Hb_109002_020 |
0.1257472438 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 16 |
Hb_002391_210 |
0.1259326594 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 17 |
Hb_007747_080 |
0.1265997624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000327_250 |
0.127049756 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 19 |
Hb_143629_100 |
0.1274857125 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 20 |
Hb_004934_050 |
0.128549967 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |