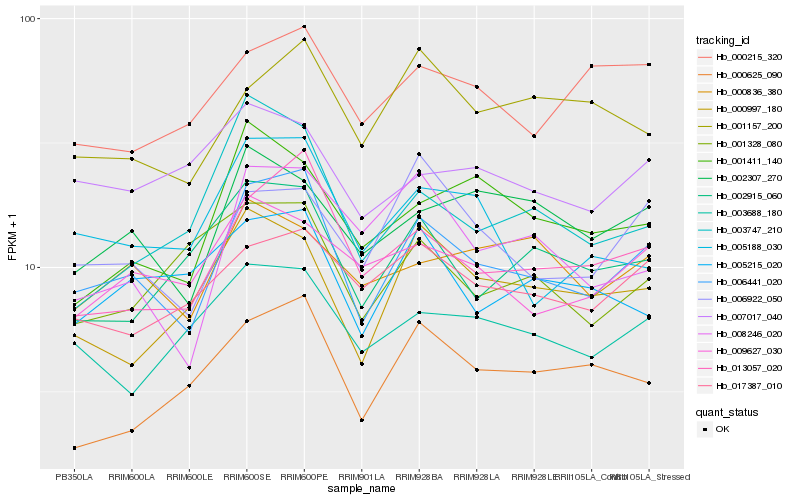
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006441_020 |
0.0 |
- |
- |
PREDICTED: cadmium/zinc-transporting ATPase HMA3-like isoform X2 [Populus euphratica] |
| 2 |
Hb_013057_020 |
0.0922682542 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 3 |
Hb_006922_050 |
0.1206570778 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 4 |
Hb_005188_030 |
0.1228337235 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_002307_270 |
0.123941406 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 6 |
Hb_017387_010 |
0.1256666104 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 7 |
Hb_002915_060 |
0.1268456732 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 8 |
Hb_008246_020 |
0.1270580328 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003688_180 |
0.127305159 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 10 |
Hb_009627_030 |
0.1273793835 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 11 |
Hb_000997_180 |
0.1276200553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005215_020 |
0.1291739047 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 13 |
Hb_001411_140 |
0.1301785354 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 14 |
Hb_003747_210 |
0.1306788937 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 15 |
Hb_001157_200 |
0.1322254009 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 16 |
Hb_000836_380 |
0.1346337693 |
- |
- |
PREDICTED: C-terminal binding protein AN-like [Populus euphratica] |
| 17 |
Hb_001328_080 |
0.1348793964 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 18 |
Hb_000215_320 |
0.1350241103 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 19 |
Hb_000625_090 |
0.1355226466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007017_040 |
0.1362097331 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |