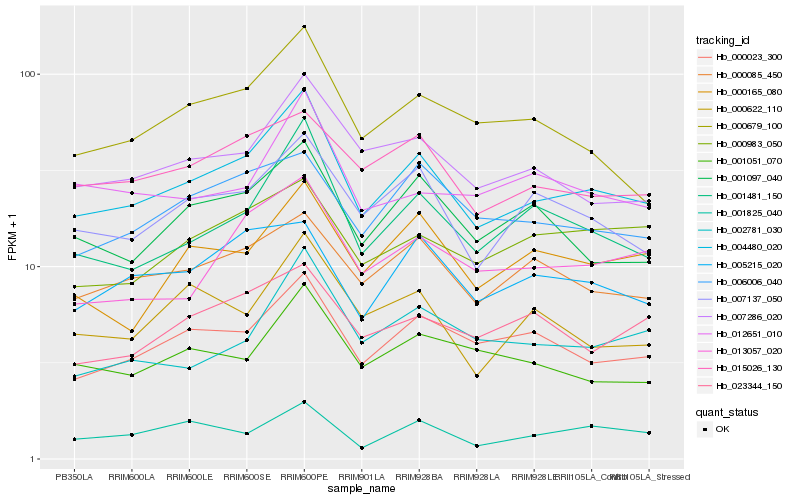
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004480_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 2 |
Hb_001481_150 |
0.0863456751 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 3 |
Hb_007286_020 |
0.0941869503 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 4 |
Hb_007137_050 |
0.0942703055 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 5 |
Hb_000023_300 |
0.0946772076 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 6 |
Hb_000085_450 |
0.1014419601 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 7 |
Hb_001097_040 |
0.1027370263 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 8 |
Hb_000679_100 |
0.1031978853 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 9 |
Hb_013057_020 |
0.1032322852 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 10 |
Hb_015026_130 |
0.104853298 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_000165_080 |
0.1101410875 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001051_070 |
0.1105062233 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_006006_040 |
0.1113392554 |
- |
- |
S-methyl-5-thioribose kinase isoform 2 [Theobroma cacao] |
| 14 |
Hb_000622_110 |
0.1113950668 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 15 |
Hb_005215_020 |
0.1132733927 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 16 |
Hb_002781_030 |
0.1148253995 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_012651_010 |
0.1150367232 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 18 |
Hb_001825_040 |
0.1152890734 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 19 |
Hb_000983_050 |
0.1168123054 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_023344_150 |
0.1178545336 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |