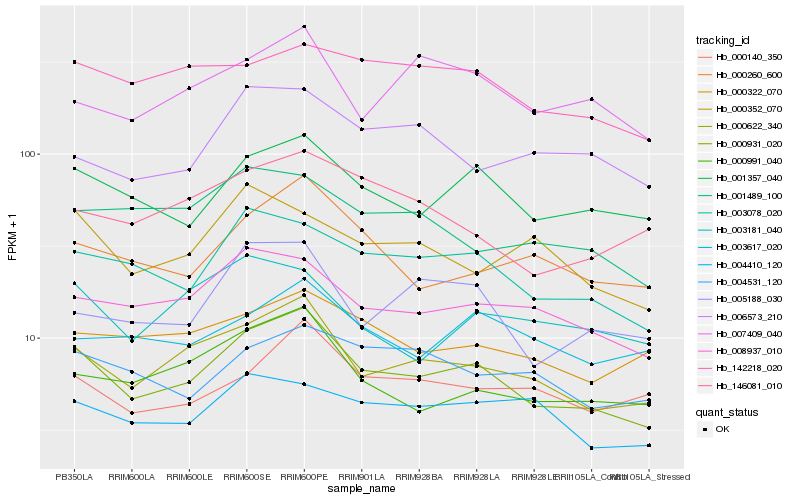
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000931_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000622_340 |
0.0773738944 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 3 |
Hb_003078_020 |
0.0963744814 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 4 |
Hb_001357_040 |
0.0979746093 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 5 |
Hb_008937_010 |
0.1061605606 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000991_040 |
0.1097423503 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 7 |
Hb_000260_600 |
0.114175329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005188_030 |
0.1154342821 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000140_350 |
0.1203304052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000322_070 |
0.1241047885 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_001489_100 |
0.1256657407 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 12 |
Hb_142218_020 |
0.1263177151 |
- |
- |
actin family protein [Populus trichocarpa] |
| 13 |
Hb_146081_010 |
0.1266107149 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 14 |
Hb_004531_120 |
0.1268040714 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 15 |
Hb_004410_120 |
0.1277749248 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007409_040 |
0.1281456671 |
- |
- |
actin family protein [Populus trichocarpa] |
| 17 |
Hb_003617_020 |
0.129613302 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 18 |
Hb_000352_070 |
0.1305563996 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 19 |
Hb_006573_210 |
0.1309221142 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 20 |
Hb_003181_040 |
0.1321781488 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |