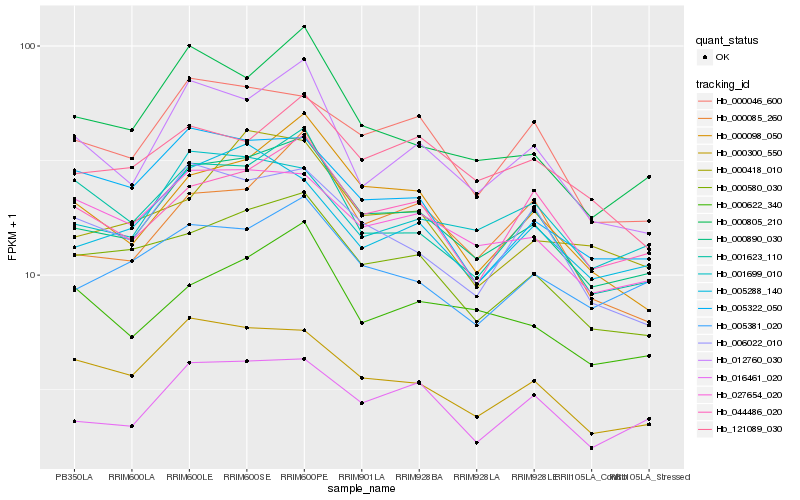
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_030 |
0.0 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 2 |
Hb_000580_030 |
0.0698409825 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 3 |
Hb_012760_030 |
0.0737457564 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 4 |
Hb_016461_020 |
0.0753499978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000098_050 |
0.0761827594 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 6 |
Hb_044486_020 |
0.0762711537 |
- |
- |
CASTOR protein [Glycine max] |
| 7 |
Hb_000805_210 |
0.0785424976 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 8 |
Hb_000300_550 |
0.0814709016 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 9 |
Hb_027654_020 |
0.0816276871 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 10 |
Hb_005381_020 |
0.0821225057 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 11 |
Hb_001623_110 |
0.0844746225 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000046_600 |
0.0855417334 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 13 |
Hb_000622_340 |
0.0859597379 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 14 |
Hb_001699_010 |
0.0882916628 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 15 |
Hb_000085_260 |
0.0893526874 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 16 |
Hb_005288_140 |
0.0905625934 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 17 |
Hb_006022_010 |
0.0912157517 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 18 |
Hb_000418_010 |
0.0930242532 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 19 |
Hb_005322_050 |
0.0940837678 |
- |
- |
hypothetical protein POPTR_0009s02400g [Populus trichocarpa] |
| 20 |
Hb_121089_030 |
0.0942901058 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |