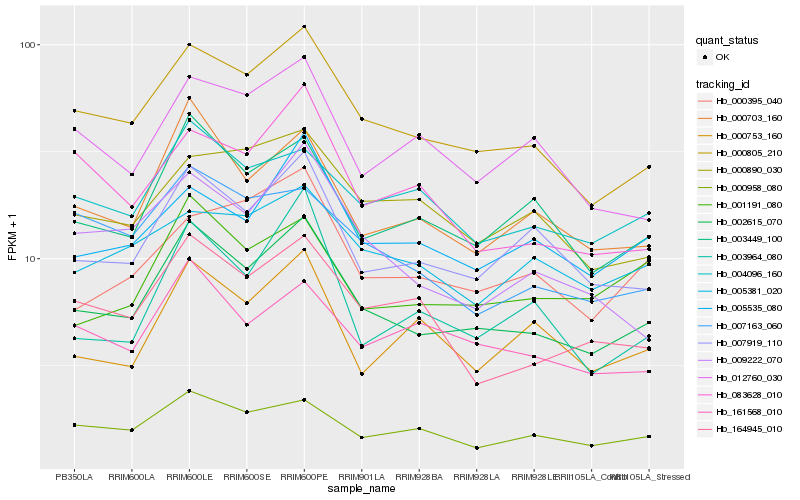
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002615_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000805_210 |
0.0608087584 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 3 |
Hb_007919_110 |
0.0872532758 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 4 |
Hb_000703_160 |
0.0877590172 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003449_100 |
0.0888922755 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 6 |
Hb_000958_080 |
0.0990987697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005535_080 |
0.1078200798 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 8 |
Hb_004096_160 |
0.1085083595 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 9 |
Hb_164945_010 |
0.1089086262 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 10 |
Hb_161568_010 |
0.1093158971 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 11 |
Hb_001191_080 |
0.1104457804 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_009222_070 |
0.1121801864 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_007163_060 |
0.115181209 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 14 |
Hb_000890_030 |
0.1162511217 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 15 |
Hb_000395_040 |
0.1167286384 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 16 |
Hb_012760_030 |
0.1170661003 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 10 [Gossypium raimondii] |
| 17 |
Hb_000753_160 |
0.1176660021 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 18 |
Hb_003964_080 |
0.1193355204 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 19 |
Hb_083628_010 |
0.1195813789 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 20 |
Hb_005381_020 |
0.1201630102 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |