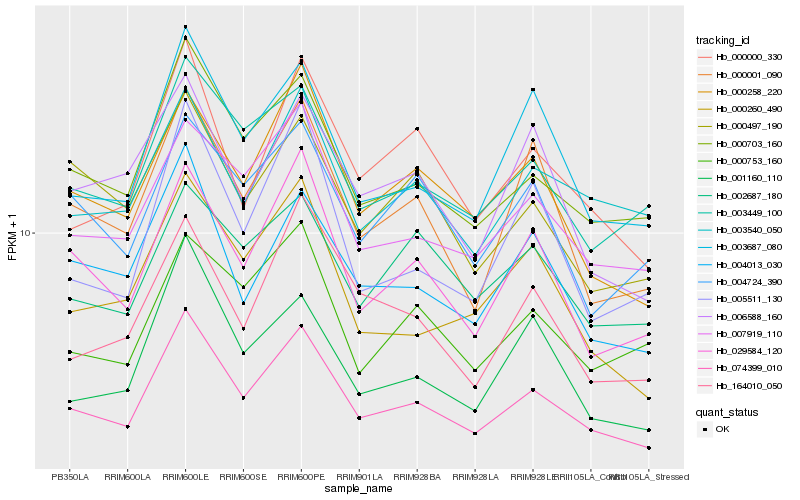
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074399_010 |
0.0 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 2 |
Hb_000703_160 |
0.0674305697 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_029584_120 |
0.080997335 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000258_220 |
0.0826708256 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003687_080 |
0.083182885 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 6 |
Hb_007919_110 |
0.0853980046 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 7 |
Hb_004013_030 |
0.0858862193 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 8 |
Hb_000000_330 |
0.086254065 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 9 |
Hb_003449_100 |
0.0876510363 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 10 |
Hb_000001_090 |
0.0877388222 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 11 |
Hb_000260_490 |
0.0945353448 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 12 |
Hb_003540_050 |
0.0971864415 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 13 |
Hb_005511_130 |
0.100781495 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 14 |
Hb_001160_110 |
0.1013803286 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 15 |
Hb_000753_160 |
0.1028785506 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 16 |
Hb_006588_160 |
0.1047284127 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_000497_190 |
0.1053070142 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 18 |
Hb_004724_390 |
0.1060402035 |
- |
- |
PREDICTED: nicalin-1-like [Populus euphratica] |
| 19 |
Hb_164010_050 |
0.1061914775 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 20 |
Hb_002687_180 |
0.1068787296 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |