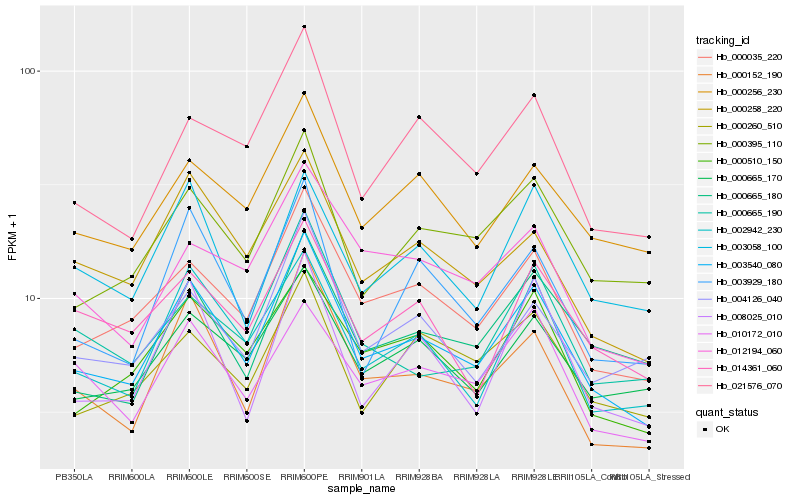
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_080 |
0.0 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000035_220 |
0.0682639722 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 3 |
Hb_000152_190 |
0.0808072108 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 4 |
Hb_002942_230 |
0.0837218627 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 5 |
Hb_000510_150 |
0.0873389641 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000258_220 |
0.0900981025 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000665_170 |
0.0906919761 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 8 |
Hb_000256_230 |
0.0912930667 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 9 |
Hb_000395_110 |
0.0928297704 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 10 |
Hb_021576_070 |
0.0956643993 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 11 |
Hb_000665_180 |
0.0982666444 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_012194_060 |
0.1001311638 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 13 |
Hb_010172_010 |
0.1002717479 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 14 |
Hb_003929_180 |
0.1010475767 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 15 |
Hb_004126_040 |
0.1012083419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003058_100 |
0.1042914671 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000260_510 |
0.1044602004 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 18 |
Hb_014361_060 |
0.1047025714 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 19 |
Hb_008025_010 |
0.1049952699 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 20 |
Hb_000665_190 |
0.1053080079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |