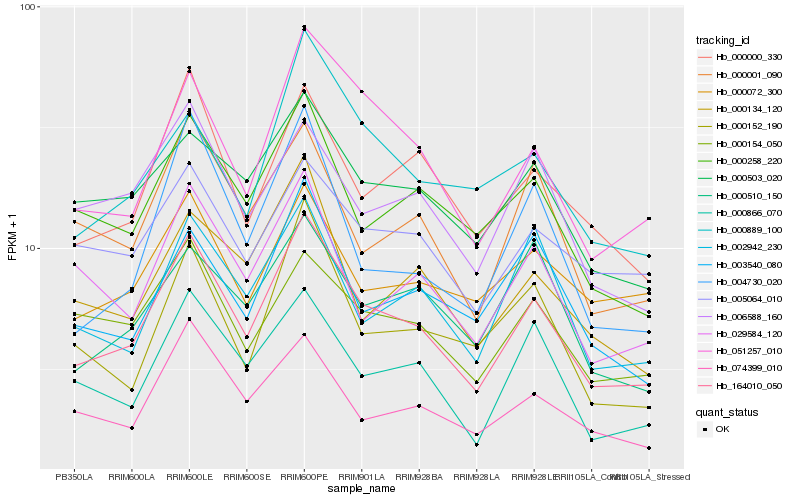
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164010_050 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 2 |
Hb_051257_010 |
0.0874462333 |
- |
- |
- |
| 3 |
Hb_000866_070 |
0.1033693879 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: SKP1-like protein 21 [Jatropha curcas] |
| 4 |
Hb_000000_330 |
0.1036542017 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 5 |
Hb_000258_220 |
0.1055535741 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_074399_010 |
0.1061914775 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 7 |
Hb_000503_020 |
0.1084090872 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 8 |
Hb_000154_050 |
0.1122814201 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 9 |
Hb_000001_090 |
0.1123255156 |
- |
- |
PREDICTED: uncharacterized protein At3g58460 [Populus euphratica] |
| 10 |
Hb_003540_080 |
0.1146697755 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000152_190 |
0.1150149138 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |
| 12 |
Hb_029584_120 |
0.1150355144 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002942_230 |
0.115122969 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 14 |
Hb_000134_120 |
0.1161030611 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 15 |
Hb_005064_010 |
0.1162807794 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 16 |
Hb_000889_100 |
0.1166885184 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 17 |
Hb_004730_020 |
0.1169579608 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 18 |
Hb_006588_160 |
0.118045209 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_000072_300 |
0.1194463893 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 20 |
Hb_000510_150 |
0.1205525903 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X1 [Jatropha curcas] |