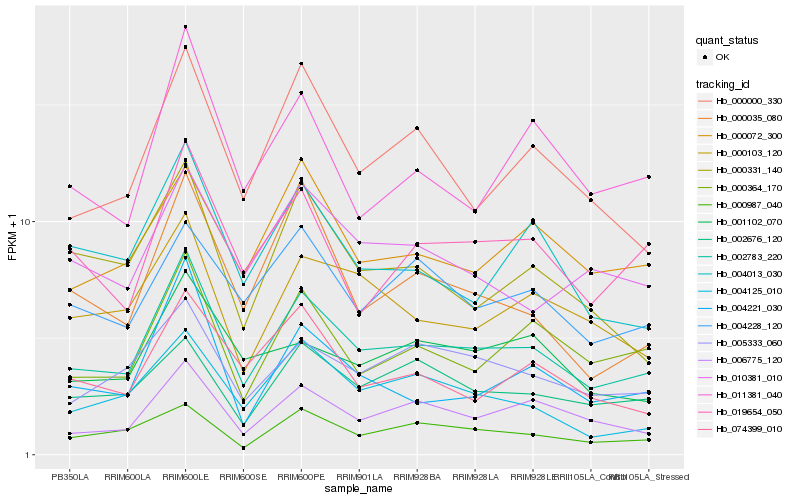
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006775_120 |
0.1022304062 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 3 |
Hb_000364_170 |
0.103151895 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 4 |
Hb_000000_330 |
0.1078960579 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 5 |
Hb_005333_060 |
0.1112489684 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 6 |
Hb_019654_050 |
0.1122780061 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 7 |
Hb_000987_040 |
0.1147220529 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 2 homolog B isoform X2 [Jatropha curcas] |
| 8 |
Hb_000103_120 |
0.1154117627 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_011381_040 |
0.1156827092 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004013_030 |
0.1156857188 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 11 |
Hb_000072_300 |
0.1174747961 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 12 |
Hb_004125_010 |
0.1201347067 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 13 |
Hb_004228_120 |
0.1210036459 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 14 |
Hb_002676_120 |
0.1215425318 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 15 |
Hb_010381_010 |
0.1216421983 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_074399_010 |
0.1217164833 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 17 |
Hb_000035_080 |
0.1217480872 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 18 |
Hb_004221_030 |
0.122431726 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000331_140 |
0.122553905 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 20 |
Hb_001102_070 |
0.1245821445 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |