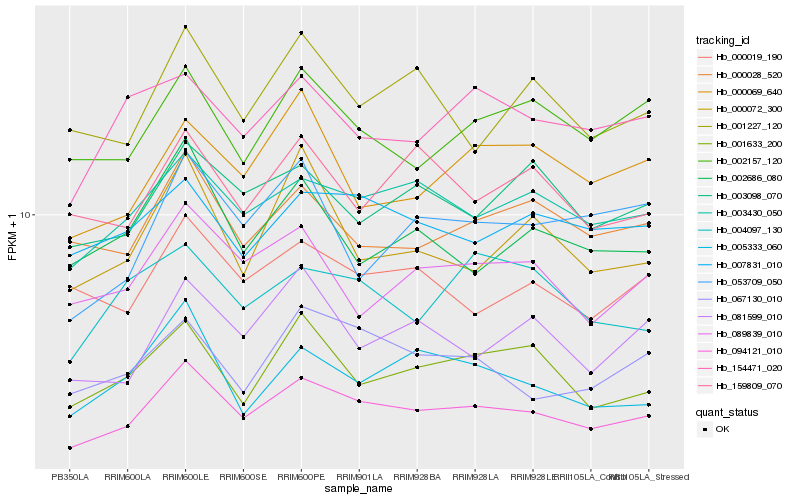
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094121_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003430_050 |
0.1025773218 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 3 |
Hb_081599_010 |
0.1101510197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001633_200 |
0.1115521775 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 5 |
Hb_000072_300 |
0.1120472185 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 6 |
Hb_067130_010 |
0.1121900898 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_005333_060 |
0.1130277044 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 8 |
Hb_089839_010 |
0.1150799441 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 9 |
Hb_001227_120 |
0.1165696227 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 10 |
Hb_053709_050 |
0.1179436941 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_002686_080 |
0.1192821454 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 12 |
Hb_000028_520 |
0.1198849838 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 13 |
Hb_000069_640 |
0.1207789387 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 14 |
Hb_003098_070 |
0.1215277031 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 15 |
Hb_002157_120 |
0.1216191696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000019_190 |
0.1224404093 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 17 |
Hb_007831_010 |
0.1254781133 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 18 |
Hb_159809_070 |
0.125751579 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 19 |
Hb_154471_020 |
0.1261716747 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 20 |
Hb_004097_130 |
0.1265049213 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |