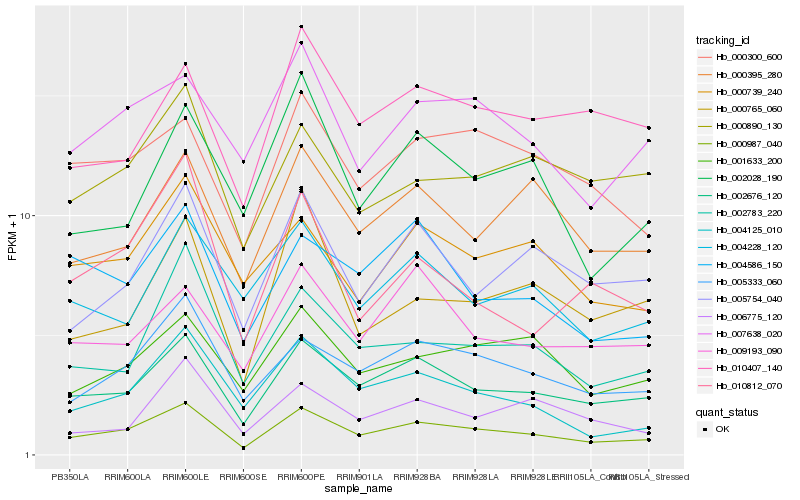
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000987_040 |
0.0 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 2 homolog B isoform X2 [Jatropha curcas] |
| 2 |
Hb_002676_120 |
0.0666091161 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 3 |
Hb_005333_060 |
0.0902330035 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 4 |
Hb_002783_220 |
0.1147220529 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005754_040 |
0.1157644118 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001633_200 |
0.1162366966 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 7 |
Hb_004125_010 |
0.1170042928 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 8 |
Hb_000395_280 |
0.1179232334 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 9 |
Hb_000300_600 |
0.1183425629 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 10 |
Hb_007638_020 |
0.1200050595 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 11 |
Hb_000739_240 |
0.1220698903 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004586_150 |
0.1252978119 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 13 |
Hb_000890_130 |
0.1280505766 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 14 |
Hb_009193_090 |
0.1286288309 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004228_120 |
0.1289168449 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 16 |
Hb_002028_190 |
0.1305480056 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 17 |
Hb_010407_140 |
0.1332968318 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 18 |
Hb_006775_120 |
0.1343543407 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 19 |
Hb_000765_060 |
0.1344259141 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 20 |
Hb_010812_070 |
0.1344750939 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |