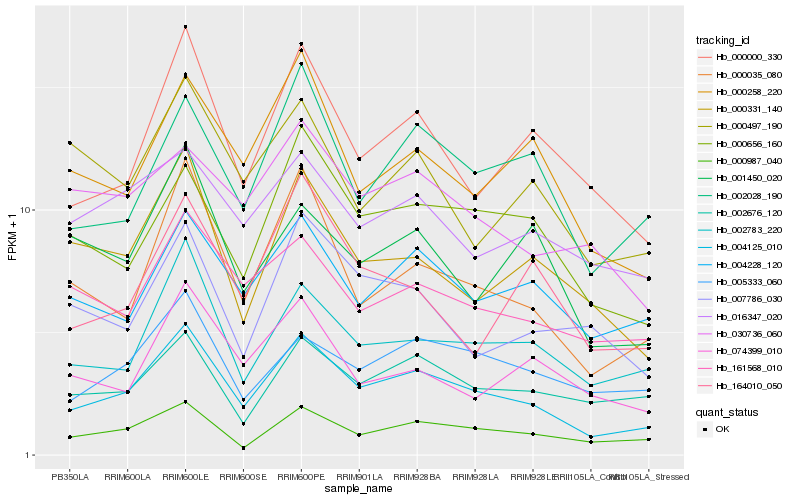
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004125_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 2 |
Hb_000035_080 |
0.106796794 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 3 |
Hb_000987_040 |
0.1170042928 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 2 homolog B isoform X2 [Jatropha curcas] |
| 4 |
Hb_002783_220 |
0.1201347067 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000000_330 |
0.1208717175 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 6 |
Hb_002028_190 |
0.123742116 |
- |
- |
PREDICTED: 26S protease regulatory subunit 7-like [Jatropha curcas] |
| 7 |
Hb_000656_160 |
0.1247489982 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 8 |
Hb_004228_120 |
0.1279887094 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 9 |
Hb_000258_220 |
0.1297252567 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005333_060 |
0.1314010344 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 11 |
Hb_161568_010 |
0.1347714451 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 12 |
Hb_000331_140 |
0.1352237489 |
- |
- |
PREDICTED: cycloeucalenol cycloisomerase [Jatropha curcas] |
| 13 |
Hb_002676_120 |
0.137808485 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 14 |
Hb_016347_020 |
0.1443967296 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 15 |
Hb_001450_020 |
0.1462424658 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_007786_030 |
0.1469170918 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 17 |
Hb_030736_060 |
0.1469708364 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 18 |
Hb_074399_010 |
0.1473834668 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 19 |
Hb_000497_190 |
0.1478955767 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 20 |
Hb_164010_050 |
0.148316286 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |