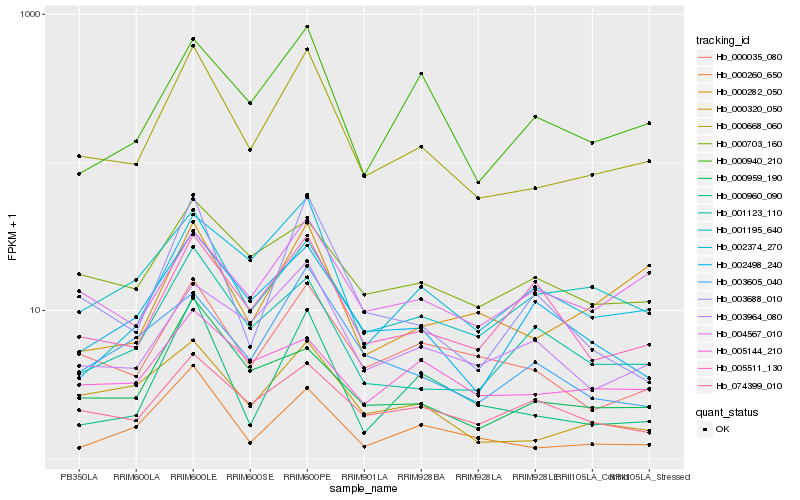
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_060 |
0.0 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 2 |
Hb_000035_080 |
0.1289664102 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 3 |
Hb_000320_050 |
0.1365775126 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 4 |
Hb_003964_080 |
0.1456513741 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 5 |
Hb_000260_650 |
0.1511768388 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 6 |
Hb_074399_010 |
0.1529176823 |
- |
- |
PREDICTED: phragmoplast orienting kinesin-1 isoform X1 [Populus euphratica] |
| 7 |
Hb_000940_210 |
0.153072114 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 8 |
Hb_000703_160 |
0.1534460298 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000959_190 |
0.1553937477 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 10 |
Hb_005511_130 |
0.1555472406 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |
| 11 |
Hb_002498_240 |
0.1559861019 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004567_010 |
0.1587430026 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 13 |
Hb_001123_110 |
0.159503543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003688_010 |
0.1600376431 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3-like [Jatropha curcas] |
| 15 |
Hb_003605_040 |
0.160670741 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002374_270 |
0.162152449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001195_640 |
0.1633071939 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 18 |
Hb_000282_050 |
0.1641112057 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 19 |
Hb_000960_090 |
0.1645138236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005144_210 |
0.1653249654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |