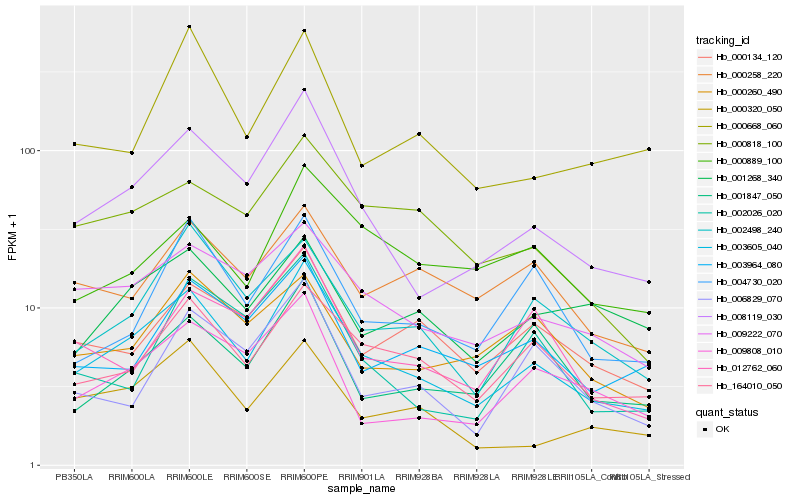
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003605_040 |
0.0 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_008119_030 |
0.0924718273 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 3 |
Hb_164010_050 |
0.1275133989 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 4 |
Hb_009222_070 |
0.1299105238 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 5 |
Hb_002498_240 |
0.1449891943 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009808_010 |
0.1475044214 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000818_100 |
0.148057176 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 8 |
Hb_000134_120 |
0.1487844331 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 9 |
Hb_000320_050 |
0.151618866 |
- |
- |
bcr-associated protein, bap, putative [Ricinus communis] |
| 10 |
Hb_003964_080 |
0.1524943012 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 11 |
Hb_004730_020 |
0.159095033 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 12 |
Hb_000668_060 |
0.160670741 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 13 |
Hb_012762_060 |
0.1612570247 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_006829_070 |
0.1637841862 |
- |
- |
PREDICTED: uncharacterized protein LOC105632920 [Jatropha curcas] |
| 15 |
Hb_001847_050 |
0.1639142359 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 16 |
Hb_000258_220 |
0.1646458983 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000260_490 |
0.1655266296 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002026_020 |
0.1665332208 |
- |
- |
hypothetical protein POPTR_0012s129501g, partial [Populus trichocarpa] |
| 19 |
Hb_001268_340 |
0.1666217527 |
- |
- |
Actin, putative [Ricinus communis] |
| 20 |
Hb_000889_100 |
0.1668845609 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |