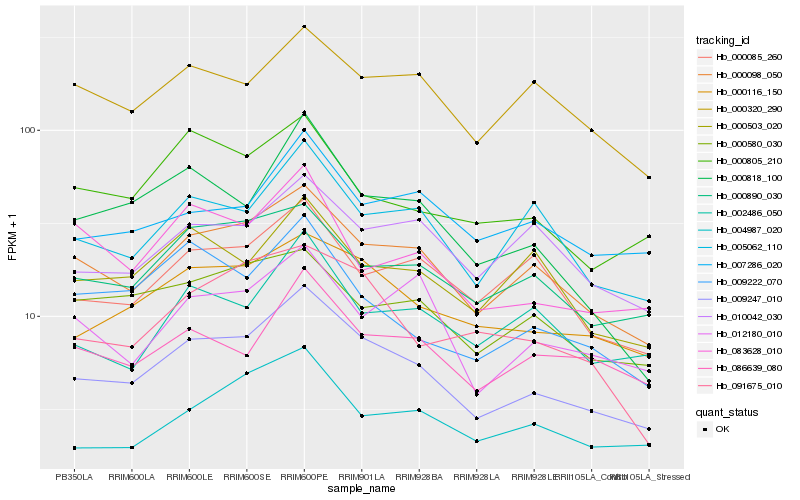
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009247_010 |
0.0 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000098_050 |
0.0736781982 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 3 |
Hb_000116_150 |
0.0880774132 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 4 |
Hb_005062_110 |
0.106917776 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 5 |
Hb_000890_030 |
0.1090907892 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 6 |
Hb_000818_100 |
0.1148782109 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 7 |
Hb_009222_070 |
0.1173115544 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_086639_080 |
0.1186014336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000085_260 |
0.1191398322 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 10 |
Hb_004987_020 |
0.1192917843 |
- |
- |
- |
| 11 |
Hb_000320_290 |
0.1194803533 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 12 |
Hb_000503_020 |
0.120788948 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 13 |
Hb_007286_020 |
0.1208782446 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 14 |
Hb_091675_010 |
0.1215910788 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_000805_210 |
0.1216208362 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 16 |
Hb_012180_010 |
0.1218278039 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |
| 17 |
Hb_000580_030 |
0.1218289716 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 18 |
Hb_083628_010 |
0.1222011564 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 19 |
Hb_002486_050 |
0.1226834795 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_010042_030 |
0.1243563467 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |