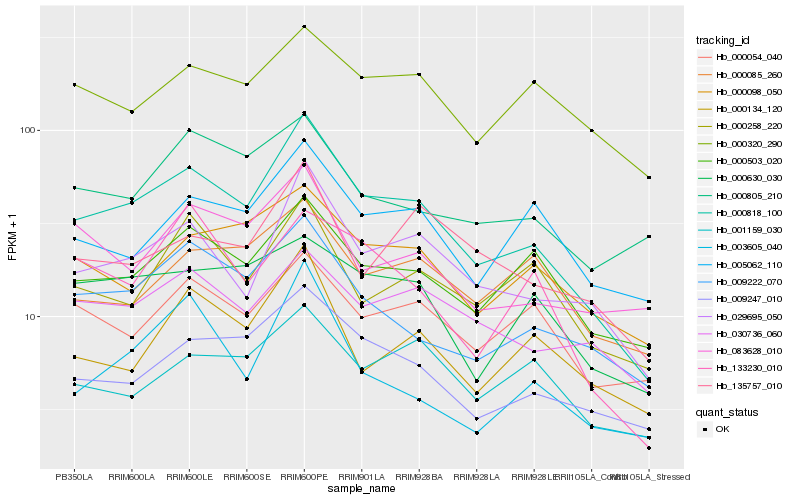
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000818_100 |
0.0 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 2 |
Hb_029695_050 |
0.1004484031 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 1 [Jatropha curcas] |
| 3 |
Hb_009247_010 |
0.1148782109 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009222_070 |
0.1220772754 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 5 |
Hb_000503_020 |
0.1302149777 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 6 |
Hb_000098_050 |
0.1395625283 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 7 |
Hb_000134_120 |
0.1410205263 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 8 |
Hb_030736_060 |
0.1414876752 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 9 |
Hb_005062_110 |
0.1417322456 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 10 |
Hb_000630_030 |
0.1421573205 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 11 |
Hb_000258_220 |
0.1434027225 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_083628_010 |
0.1448435265 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 13 |
Hb_000054_040 |
0.1469216805 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 14 |
Hb_135757_010 |
0.1471688365 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 15 |
Hb_133230_010 |
0.148047336 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 16 |
Hb_003605_040 |
0.148057176 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001159_030 |
0.149064009 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 18 |
Hb_000805_210 |
0.1528987534 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 19 |
Hb_000085_260 |
0.1534919597 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_000320_290 |
0.1548538911 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |