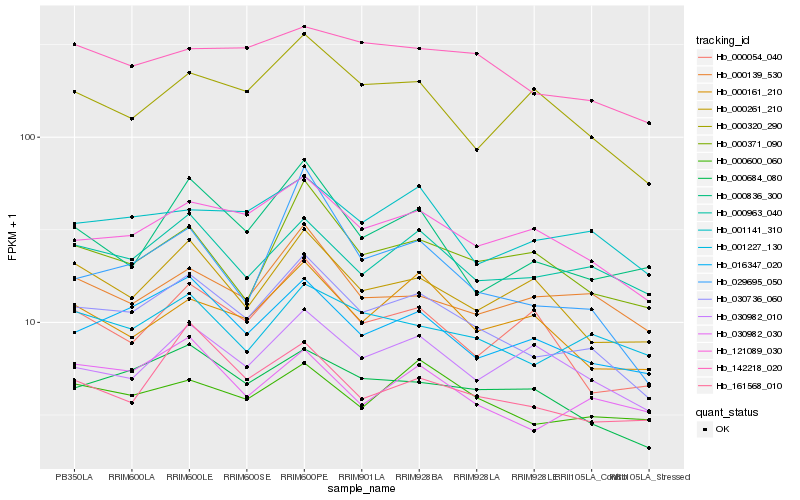
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030736_060 |
0.0 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 2 |
Hb_016347_020 |
0.0896051323 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000684_080 |
0.090800985 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 4 |
Hb_121089_030 |
0.0913858521 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 5 |
Hb_000963_040 |
0.1039833843 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 6 |
Hb_000261_210 |
0.1058461302 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_001227_130 |
0.1061904175 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 8 |
Hb_030982_030 |
0.1081409378 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000320_290 |
0.1085319916 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 10 |
Hb_161568_010 |
0.1089598977 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 11 |
Hb_000371_090 |
0.1092256318 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 12 |
Hb_142218_020 |
0.1103157119 |
- |
- |
actin family protein [Populus trichocarpa] |
| 13 |
Hb_000600_060 |
0.1104056618 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001141_310 |
0.1113038177 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 15 |
Hb_000161_210 |
0.1114529848 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 16 |
Hb_030982_010 |
0.1115567416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000836_300 |
0.1132814628 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 18 |
Hb_000054_040 |
0.1149041692 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 19 |
Hb_000139_530 |
0.114948723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_029695_050 |
0.1171222563 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 1 [Jatropha curcas] |