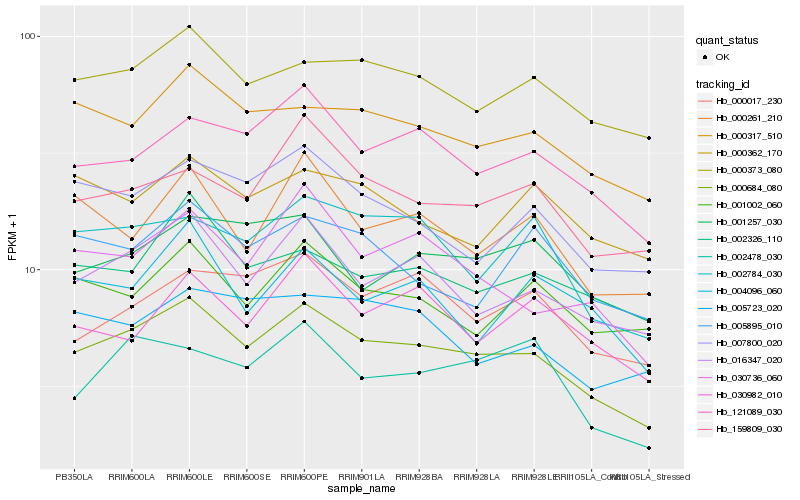
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_080 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 2 |
Hb_016347_020 |
0.0744691623 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 3 |
Hb_121089_030 |
0.0814503749 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 4 |
Hb_000317_510 |
0.0845257338 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 5 |
Hb_000373_080 |
0.0854215076 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_030736_060 |
0.090800985 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 7 |
Hb_002784_030 |
0.0929049053 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 8 |
Hb_007800_020 |
0.0934615925 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_159809_030 |
0.0943667951 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002326_110 |
0.0979231459 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 11 |
Hb_001257_030 |
0.0996955401 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 12 |
Hb_000261_210 |
0.0997412876 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 13 |
Hb_000017_230 |
0.1000840661 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 14 |
Hb_001002_060 |
0.1002361819 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 15 |
Hb_004096_060 |
0.1036997179 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 16 |
Hb_005723_020 |
0.1045038258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005895_010 |
0.1048015461 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002478_030 |
0.1059024733 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 19 |
Hb_000362_170 |
0.1060230999 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 20 |
Hb_030982_010 |
0.106039303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |