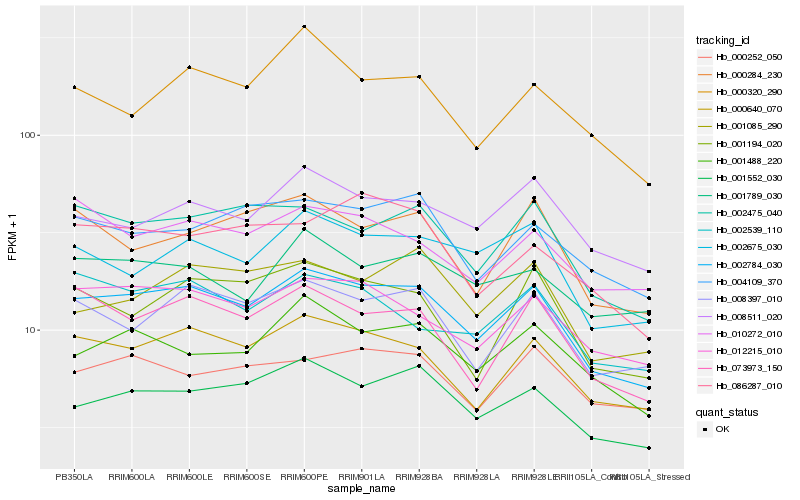
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002784_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 2 |
Hb_000640_070 |
0.0582312293 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 3 |
Hb_001552_030 |
0.0679524296 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_012215_010 |
0.0703107235 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 5 |
Hb_008397_010 |
0.0718056972 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002475_040 |
0.0735219238 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_073973_150 |
0.0757899148 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001488_220 |
0.079279449 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 9 |
Hb_002675_030 |
0.0806974388 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 10 |
Hb_008511_020 |
0.0817182579 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 11 |
Hb_004109_370 |
0.0837639735 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_001194_020 |
0.0839332226 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 13 |
Hb_000284_230 |
0.0863020027 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 14 |
Hb_002539_110 |
0.0876761531 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 15 |
Hb_010272_010 |
0.0880609011 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 16 |
Hb_001085_290 |
0.0885075898 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 17 |
Hb_000252_050 |
0.0890574132 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 18 |
Hb_000320_290 |
0.0894406626 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 19 |
Hb_001789_030 |
0.0923187177 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 20 |
Hb_086287_010 |
0.0926503178 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |