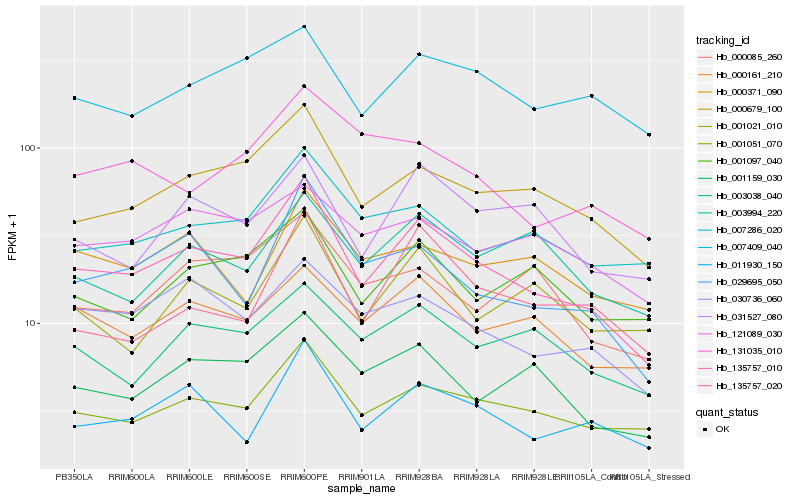
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135757_010 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 2 |
Hb_000679_100 |
0.0963636446 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001051_070 |
0.0990857227 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_029695_050 |
0.1111682815 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 1 [Jatropha curcas] |
| 5 |
Hb_011930_150 |
0.1121695939 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001159_030 |
0.1172180394 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 7 |
Hb_030736_060 |
0.1179145913 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 8 |
Hb_000161_210 |
0.1210584251 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 9 |
Hb_007286_020 |
0.1236182797 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 10 |
Hb_031527_080 |
0.1253108503 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001097_040 |
0.1258933932 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 12 |
Hb_003038_040 |
0.1271316494 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 13 |
Hb_007409_040 |
0.1295636191 |
- |
- |
actin family protein [Populus trichocarpa] |
| 14 |
Hb_001021_010 |
0.1316254552 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 15 |
Hb_000085_260 |
0.1360526972 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 16 |
Hb_121089_030 |
0.1362787295 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 17 |
Hb_003994_220 |
0.136422149 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 18 |
Hb_135757_020 |
0.1370085553 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 19 |
Hb_000371_090 |
0.1381779257 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 20 |
Hb_131035_010 |
0.1406468342 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |