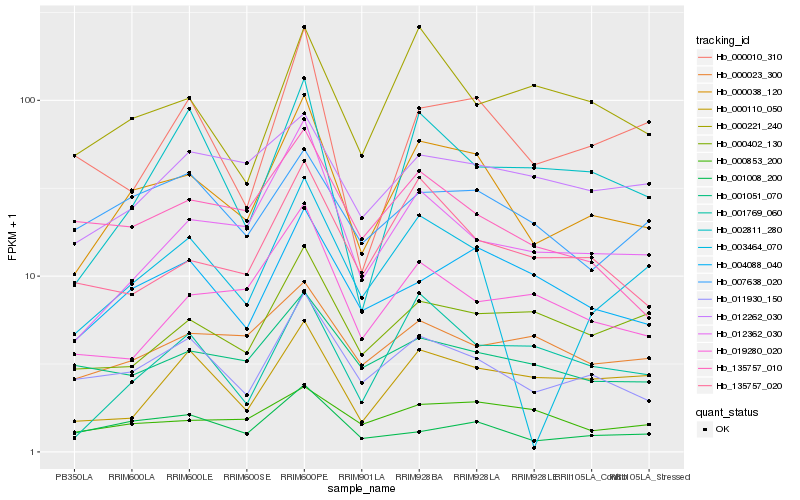
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000038_120 |
0.0 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 2 |
Hb_011930_150 |
0.1186182229 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012362_030 |
0.1432811585 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 4 |
Hb_004088_040 |
0.1464513081 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 5 |
Hb_135757_020 |
0.1488128249 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 6 |
Hb_001008_200 |
0.1559926706 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 7 |
Hb_001051_070 |
0.1599027847 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_135757_010 |
0.1615416651 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 9 |
Hb_019280_020 |
0.1628655787 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003464_070 |
0.1635937678 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 11 |
Hb_000010_310 |
0.1671698112 |
- |
- |
PREDICTED: probable pectate lyase 18 [Populus euphratica] |
| 12 |
Hb_000023_300 |
0.1671846914 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 13 |
Hb_000402_130 |
0.1680820715 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 14 |
Hb_012262_030 |
0.1693428673 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001769_060 |
0.169444781 |
- |
- |
PREDICTED: F-box protein At3g12350 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_280 |
0.1695371187 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 17 |
Hb_007638_020 |
0.1702778646 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 18 |
Hb_000853_200 |
0.1709201568 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 19 |
Hb_000221_240 |
0.1711949912 |
- |
- |
chloroplast 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_000110_050 |
0.1717095698 |
- |
- |
- |