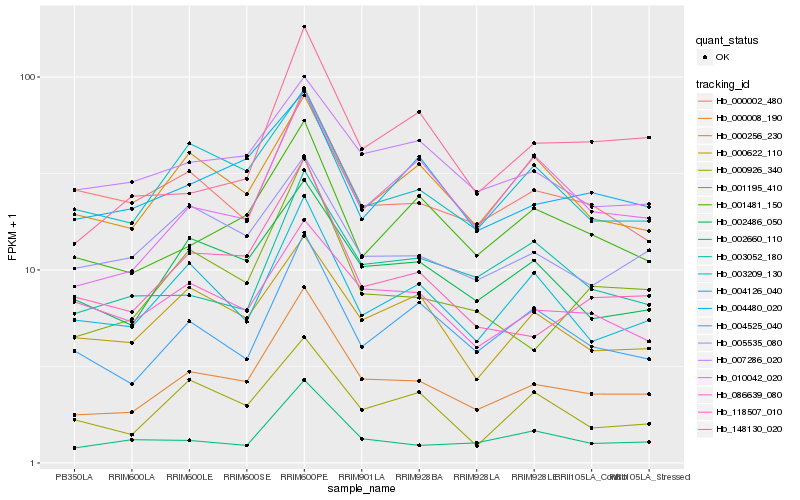
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_190 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_118507_010 |
0.1001646242 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 3 |
Hb_002486_050 |
0.1052784655 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_001195_410 |
0.1083211432 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 5 |
Hb_004126_040 |
0.1097303589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002660_110 |
0.1117301909 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001481_150 |
0.1127579309 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 8 |
Hb_004525_040 |
0.1131322837 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 9 |
Hb_003209_130 |
0.1140912601 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_000622_110 |
0.1214323397 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 11 |
Hb_004480_020 |
0.1243792348 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 12 |
Hb_005535_080 |
0.1246833642 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 13 |
Hb_000002_480 |
0.1250280479 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007286_020 |
0.1250903263 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 15 |
Hb_000926_340 |
0.125951936 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_148130_020 |
0.1268626416 |
- |
- |
- |
| 17 |
Hb_010042_020 |
0.1268734517 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 18 |
Hb_000256_230 |
0.1271855812 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 19 |
Hb_086639_080 |
0.1289877136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003052_180 |
0.130318733 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |