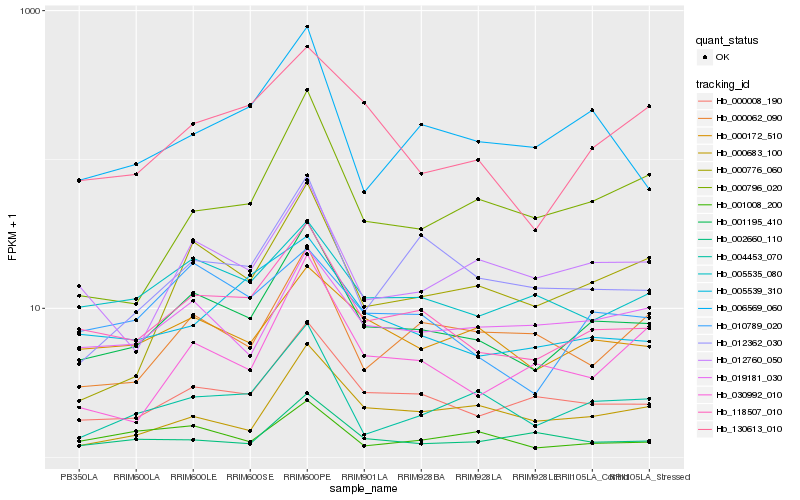
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_410 |
0.0 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 2 |
Hb_118507_010 |
0.0830109969 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 3 |
Hb_000008_190 |
0.1083211432 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004453_070 |
0.136391616 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_012760_050 |
0.1413703156 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 6 |
Hb_000172_510 |
0.1439614993 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 7 |
Hb_000776_060 |
0.1482983532 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_019181_030 |
0.1496806622 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 9 |
Hb_130613_010 |
0.1505012826 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 10 |
Hb_012362_030 |
0.1538274712 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 11 |
Hb_005535_080 |
0.1565168095 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 12 |
Hb_000683_100 |
0.1571183799 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 13 |
Hb_006569_060 |
0.1571919102 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 14 |
Hb_000062_090 |
0.1590922448 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001008_200 |
0.1607655082 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 16 |
Hb_030992_010 |
0.1611414351 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_010789_020 |
0.1611484907 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 18 |
Hb_000796_020 |
0.1614409612 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 19 |
Hb_005539_310 |
0.1621142493 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 20 |
Hb_002660_110 |
0.1630975636 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |