| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
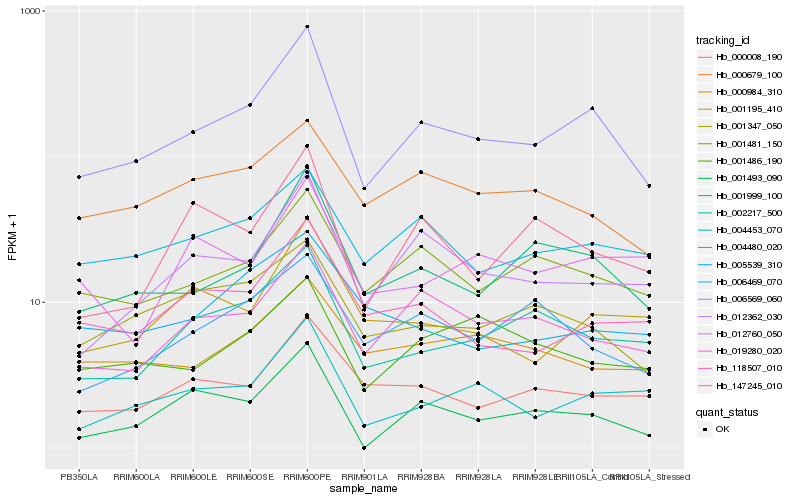
Hb_006569_060 |
0.0 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 2 |
Hb_001999_100 |
0.1162200035 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 3 |
Hb_019280_020 |
0.1374165976 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 4 |
Hb_012362_030 |
0.1378008557 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 5 |
Hb_001481_150 |
0.1384640075 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 6 |
Hb_002217_500 |
0.1436196513 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 7 |
Hb_004453_070 |
0.1457196811 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004480_020 |
0.1512020426 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 9 |
Hb_000008_190 |
0.1540861373 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_118507_010 |
0.1555087353 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 11 |
Hb_001347_050 |
0.1556852621 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001195_410 |
0.1571919102 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 13 |
Hb_147245_010 |
0.1649809013 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 14 |
Hb_000679_100 |
0.1659394304 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000984_310 |
0.1676305879 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 16 |
Hb_001486_190 |
0.1682891243 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 17 |
Hb_006469_070 |
0.1683316354 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 18 |
Hb_005539_310 |
0.1774796889 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001493_090 |
0.1777478061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012760_050 |
0.178379102 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |