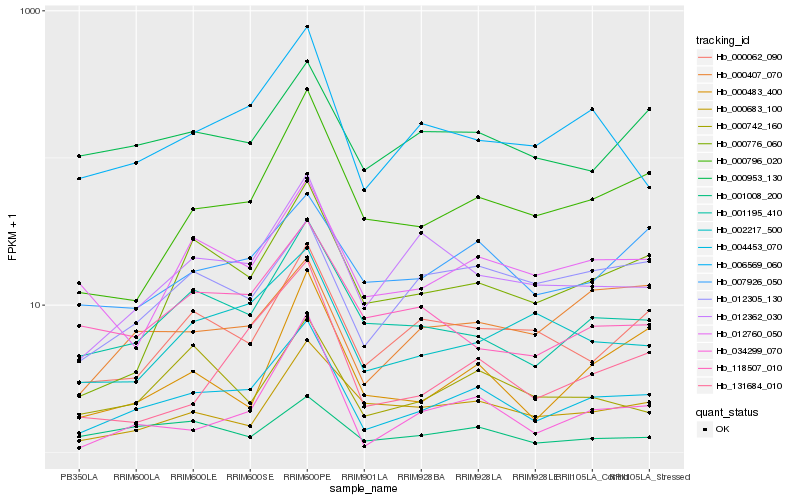
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004453_070 |
0.0 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001195_410 |
0.136391616 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 3 |
Hb_000796_020 |
0.1451924468 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 4 |
Hb_006569_060 |
0.1457196811 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 5 |
Hb_000776_060 |
0.1584714481 |
- |
- |
histone H4 [Zea mays] |
| 6 |
Hb_012362_030 |
0.1586366471 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 7 |
Hb_000062_090 |
0.1606837098 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012760_050 |
0.1607237864 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 9 |
Hb_002217_500 |
0.1655491676 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 10 |
Hb_007926_050 |
0.1718226976 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 11 |
Hb_131684_010 |
0.1742675266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001008_200 |
0.175048995 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103961552 [Pyrus x bretschneideri] |
| 13 |
Hb_000683_100 |
0.1753266281 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 14 |
Hb_034299_070 |
0.1781828656 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 15 |
Hb_118507_010 |
0.1798425925 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 16 |
Hb_012305_130 |
0.1803472606 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 17 |
Hb_000953_130 |
0.1803748842 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 18 |
Hb_000742_160 |
0.1818300557 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 19 |
Hb_000483_400 |
0.1824914854 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000407_070 |
0.1842845389 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |