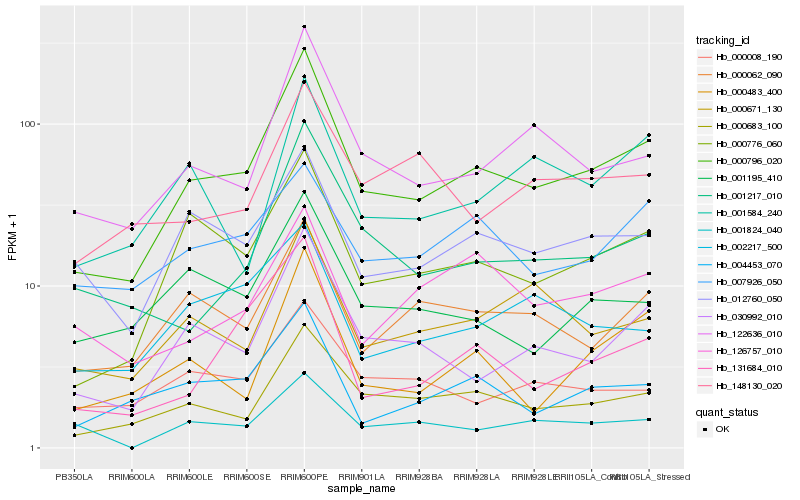
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_020 |
0.0 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 2 |
Hb_000683_100 |
0.1207921859 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 3 |
Hb_000776_060 |
0.121491399 |
- |
- |
histone H4 [Zea mays] |
| 4 |
Hb_030992_010 |
0.12494488 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 5 |
Hb_122636_010 |
0.1350487399 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 6 |
Hb_004453_070 |
0.1451924468 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000062_090 |
0.1483490438 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001217_010 |
0.1496132128 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 9 |
Hb_000483_400 |
0.1527682499 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000671_130 |
0.1531016371 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012760_050 |
0.1541985387 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 12 |
Hb_131684_010 |
0.1576631952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001195_410 |
0.1614409612 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 14 |
Hb_000008_190 |
0.1690873199 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001584_240 |
0.1695363627 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 16 |
Hb_002217_500 |
0.1712634979 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 17 |
Hb_007926_050 |
0.175723026 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 18 |
Hb_148130_020 |
0.1757289341 |
- |
- |
- |
| 19 |
Hb_126757_010 |
0.1787005421 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 20 |
Hb_001824_040 |
0.1811065311 |
- |
- |
hypothetical protein M569_11359, partial [Genlisea aurea] |