| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
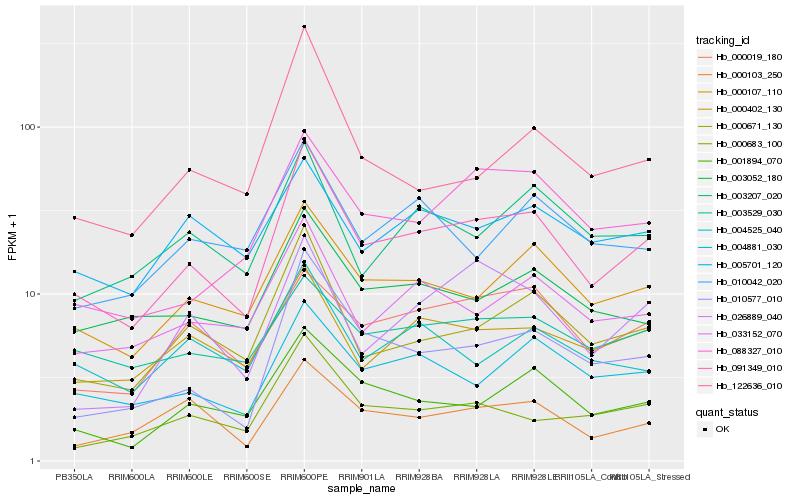
Hb_091349_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 2 |
Hb_000671_130 |
0.1092618968 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_010577_010 |
0.110933562 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 4 |
Hb_000107_110 |
0.111521068 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_001894_070 |
0.1199226766 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_033152_070 |
0.1241611032 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 7 |
Hb_004881_030 |
0.1268024349 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003052_180 |
0.1332962361 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 9 |
Hb_000402_130 |
0.1338031092 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 10 |
Hb_000683_100 |
0.1343125951 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 11 |
Hb_000103_250 |
0.1364055388 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 12 |
Hb_003207_020 |
0.1393152791 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 13 |
Hb_088327_010 |
0.1406939326 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 14 |
Hb_026889_040 |
0.1444825912 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 15 |
Hb_000019_180 |
0.1447558359 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 16 |
Hb_122636_010 |
0.1456098989 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 17 |
Hb_010042_020 |
0.1458160234 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 18 |
Hb_004525_040 |
0.1476198767 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 19 |
Hb_005701_120 |
0.1489783023 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 20 |
Hb_003529_030 |
0.150664512 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |