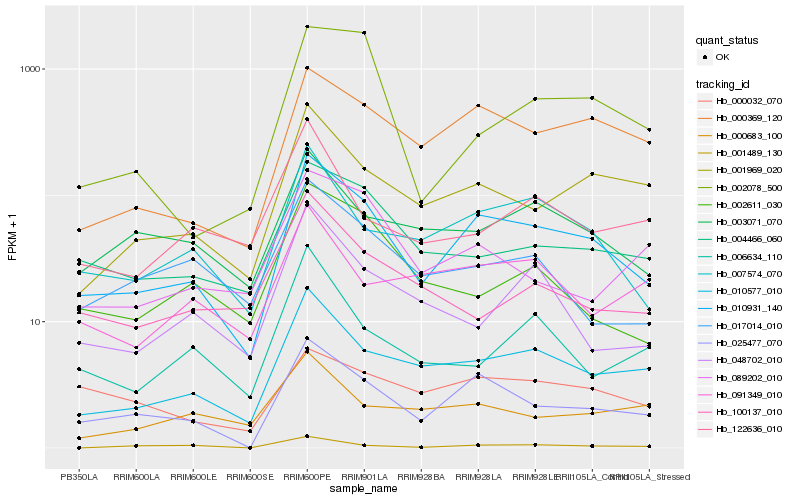
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_025477_070 |
0.1142390068 |
- |
- |
hypothetical protein CICLE_v10006100mg [Citrus clementina] |
| 3 |
Hb_010577_010 |
0.1399524603 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 4 |
Hb_007574_070 |
0.1470223701 |
- |
- |
fatty acid desaturase, putative [Ricinus communis] |
| 5 |
Hb_000369_120 |
0.1487121609 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3-B [Jatropha curcas] |
| 6 |
Hb_001969_020 |
0.1600881522 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 7 |
Hb_004466_060 |
0.1683071464 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |
| 8 |
Hb_003071_070 |
0.1788301857 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 9 |
Hb_017014_010 |
0.193278719 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002611_030 |
0.1944995073 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 11 |
Hb_089202_010 |
0.2019827004 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 12 |
Hb_000032_070 |
0.2020005316 |
- |
- |
hypothetical protein POPTR_0003s13760g [Populus trichocarpa] |
| 13 |
Hb_048702_010 |
0.2027589082 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_091349_010 |
0.2053802406 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 15 |
Hb_100137_010 |
0.2057518858 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 16 |
Hb_006634_110 |
0.2082411159 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 17 |
Hb_002078_500 |
0.2088972527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_122636_010 |
0.2094814903 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 19 |
Hb_000683_100 |
0.2096023772 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 20 |
Hb_001489_130 |
0.2112491681 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |