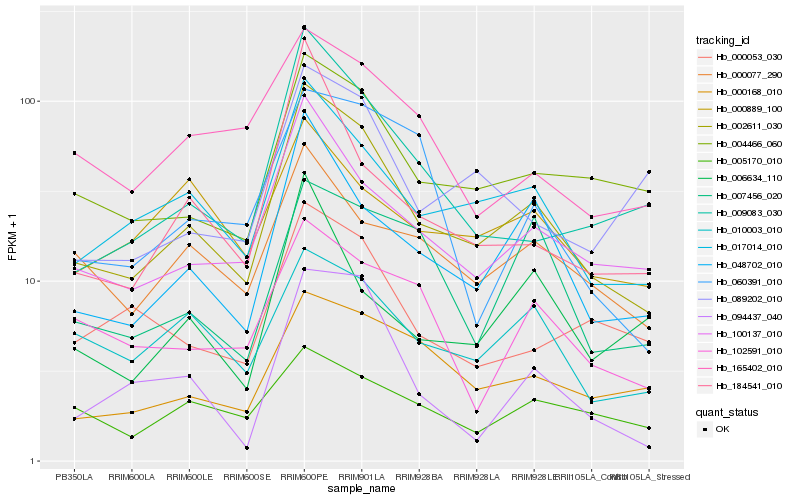
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002611_030 |
0.0 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 2 |
Hb_017014_010 |
0.1122045768 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_100137_010 |
0.1277214038 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 4 |
Hb_004466_060 |
0.1345969403 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |
| 5 |
Hb_048702_010 |
0.1347267388 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_009083_030 |
0.1514059985 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 7 |
Hb_010003_010 |
0.1630792085 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000168_010 |
0.1645850465 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000077_290 |
0.1652264797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_165402_010 |
0.1695490672 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 11 |
Hb_000053_030 |
0.1695580956 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 12 |
Hb_094437_040 |
0.1712528774 |
- |
- |
ankyrin repeat family protein, partial [Manihot esculenta] |
| 13 |
Hb_102591_010 |
0.1756337819 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 14 |
Hb_089202_010 |
0.1767701532 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Nicotiana sylvestris] |
| 15 |
Hb_184541_010 |
0.1848113787 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 16 |
Hb_000889_100 |
0.1851487046 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 17 |
Hb_007456_020 |
0.1860819564 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_060391_010 |
0.1885479245 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_005170_010 |
0.1905499422 |
- |
- |
- |
| 20 |
Hb_006634_110 |
0.1920851764 |
- |
- |
BnaC04g40780D [Brassica napus] |