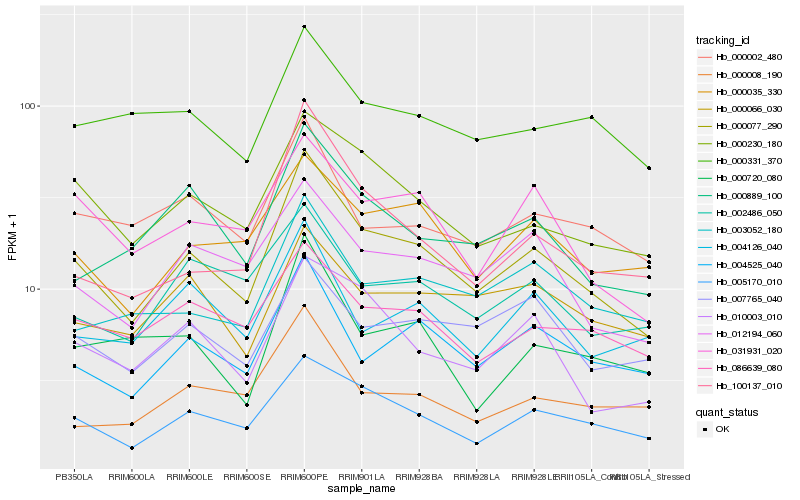
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005170_010 |
0.0953984562 |
- |
- |
- |
| 3 |
Hb_000230_180 |
0.1108152298 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_004525_040 |
0.1151925109 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 5 |
Hb_012194_060 |
0.1196160174 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 6 |
Hb_000035_330 |
0.124405722 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 7 |
Hb_000889_100 |
0.1249276185 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 8 |
Hb_010003_010 |
0.1255524555 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 9 |
Hb_031931_020 |
0.1270720602 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 10 |
Hb_086639_080 |
0.1294865852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000002_480 |
0.129828795 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003052_180 |
0.1305479461 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 13 |
Hb_100137_010 |
0.1314251781 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 14 |
Hb_002486_050 |
0.1356133281 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_004126_040 |
0.1369085928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000008_190 |
0.1373155501 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007765_040 |
0.1373574338 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 18 |
Hb_000331_370 |
0.1383866655 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 19 |
Hb_000066_030 |
0.1390368846 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000720_080 |
0.1406629603 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |