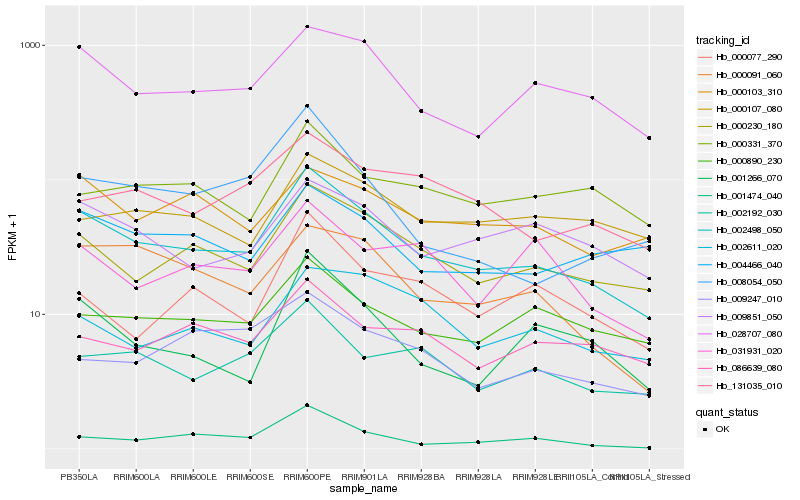
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_050 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 2 |
Hb_000230_180 |
0.1052804533 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_028707_080 |
0.1193178541 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 4 |
Hb_002192_030 |
0.1371615855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_009851_050 |
0.1403356173 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 6 |
Hb_000091_060 |
0.1404097449 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 7 |
Hb_000890_230 |
0.1427824438 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 8 |
Hb_004466_040 |
0.1476958999 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 9 |
Hb_001266_070 |
0.1485592644 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 10 |
Hb_131035_010 |
0.1487381111 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000103_310 |
0.1532676659 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 12 |
Hb_000077_290 |
0.1554101558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_086639_080 |
0.1555092795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000331_370 |
0.1555428342 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 15 |
Hb_001474_040 |
0.1563566665 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 16 |
Hb_008054_050 |
0.1571234582 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 17 |
Hb_000107_080 |
0.1571897033 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 18 |
Hb_031931_020 |
0.1574968128 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 19 |
Hb_002611_020 |
0.1592548488 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 20 |
Hb_009247_010 |
0.1593111726 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |