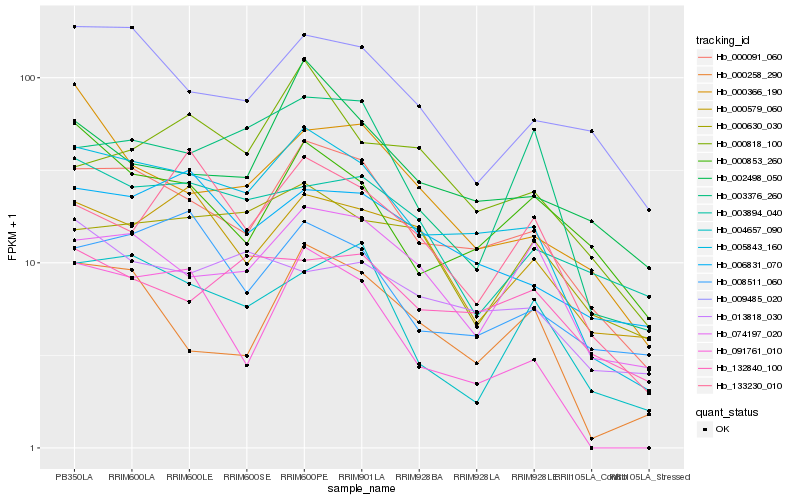
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005843_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 2 |
Hb_000091_060 |
0.0829055762 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 3 |
Hb_006831_070 |
0.1474142792 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_091761_010 |
0.1577695503 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 5 |
Hb_008511_060 |
0.1589004992 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_074197_020 |
0.1589812707 |
- |
- |
PREDICTED: uncharacterized protein LOC105645948 [Jatropha curcas] |
| 7 |
Hb_000258_290 |
0.1618415235 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 8 |
Hb_009485_020 |
0.1618490496 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 9 |
Hb_013818_030 |
0.1640697844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_132840_100 |
0.1642059613 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 11 |
Hb_000579_060 |
0.164505113 |
- |
- |
PREDICTED: uncharacterized protein LOC105633849 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000853_260 |
0.1678458777 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 13 |
Hb_003376_260 |
0.1681586716 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004657_090 |
0.1682315049 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 15 |
Hb_002498_050 |
0.1698629156 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 16 |
Hb_000366_190 |
0.1709289334 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 17 |
Hb_000818_100 |
0.1735149839 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 18 |
Hb_133230_010 |
0.1736885741 |
- |
- |
hypothetical protein L484_001846 [Morus notabilis] |
| 19 |
Hb_000630_030 |
0.1737228305 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 20 |
Hb_003894_040 |
0.1765439965 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |