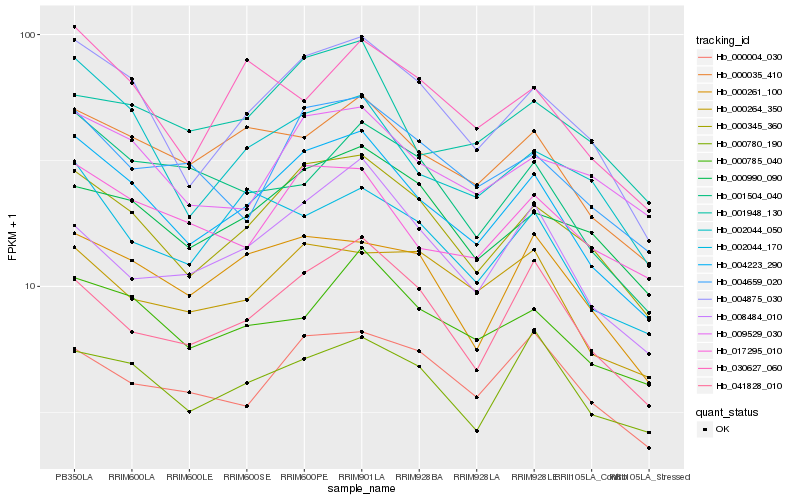
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_290 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 2 |
Hb_004875_030 |
0.0534605675 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 3 |
Hb_000345_360 |
0.0587521378 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 4 |
Hb_000035_410 |
0.079401584 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 5 |
Hb_002044_050 |
0.0855499475 |
- |
- |
- |
| 6 |
Hb_000785_040 |
0.0867784815 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 7 |
Hb_041828_010 |
0.0885849153 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 8 |
Hb_017295_010 |
0.0907561009 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 9 |
Hb_000990_090 |
0.0910041476 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_000261_100 |
0.0925123694 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000780_190 |
0.0935039503 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 12 |
Hb_004659_020 |
0.0941592254 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 13 |
Hb_002044_170 |
0.0950114946 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 14 |
Hb_009529_030 |
0.097059145 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 15 |
Hb_030627_060 |
0.0978693711 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_008484_010 |
0.0985713551 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 17 |
Hb_001948_130 |
0.0988236 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 18 |
Hb_000004_030 |
0.0991822754 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 19 |
Hb_000264_350 |
0.0994524157 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 20 |
Hb_001504_040 |
0.1008194336 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |