| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
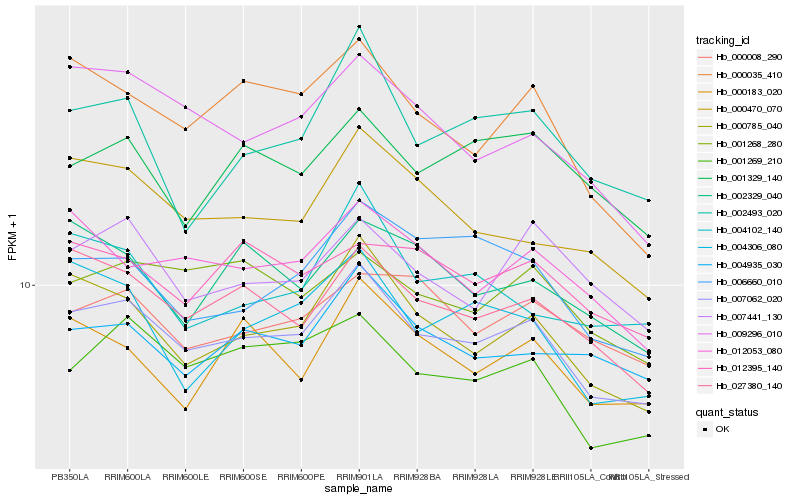
Hb_007062_020 |
0.0 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 2 |
Hb_000785_040 |
0.0546959518 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 3 |
Hb_027380_140 |
0.0600550839 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001269_210 |
0.0626733823 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000470_070 |
0.0695125028 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 6 |
Hb_009296_010 |
0.069872246 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 7 |
Hb_012053_080 |
0.0720054211 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 8 |
Hb_004935_030 |
0.0722505039 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 9 |
Hb_000035_410 |
0.0723860359 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 10 |
Hb_001268_280 |
0.0736566529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007441_130 |
0.0755003868 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000008_290 |
0.0773793924 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 13 |
Hb_006660_010 |
0.0777820605 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 14 |
Hb_004306_080 |
0.0780342388 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 15 |
Hb_012395_140 |
0.0790519909 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 16 |
Hb_001329_140 |
0.0794138052 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 17 |
Hb_000183_020 |
0.0798199334 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002329_040 |
0.0808385494 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 19 |
Hb_004102_140 |
0.0809873156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002493_020 |
0.081109946 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |